Beige Adipocyte Maintenance Is Regulated by Autophagy-Induced Mitochondrial Clearance
- PMID: 27568548
- PMCID: PMC5023491
- DOI: 10.1016/j.cmet.2016.08.002
Beige Adipocyte Maintenance Is Regulated by Autophagy-Induced Mitochondrial Clearance
Abstract
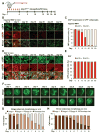
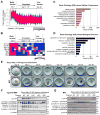
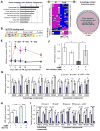
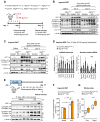
Beige adipocytes gained much attention as an alternative cellular target in anti-obesity therapy. While recent studies have identified a number of regulatory circuits that promote beige adipocyte differentiation, the molecular basis of beige adipocyte maintenance remains unknown. Here, we demonstrate that beige adipocytes progressively lose their morphological and molecular characteristics after withdrawing external stimuli and directly acquire white-like characteristics bypassing an intermediate precursor stage. The beige-to-white adipocyte transition is tightly coupled to a decrease in mitochondria, increase in autophagy, and activation of MiT/TFE transcription factor-mediated lysosome biogenesis. The autophagy pathway is crucial for mitochondrial clearance during the transition; inhibiting autophagy by uncoupled protein 1 (UCP1(+))-adipocyte-specific deletion of Atg5 or Atg12 prevents beige adipocyte loss after withdrawing external stimuli, maintaining high thermogenic capacity and protecting against diet-induced obesity and insulin resistance. The present study uncovers a fundamental mechanism by which autophagy-mediated mitochondrial clearance controls beige adipocyte maintenance, thereby providing new opportunities to counteract obesity.
Keywords: beige adipocytes; diabetes; mitochondria; mitophagy; obesity.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Metabolism: Mitophagy turns beige adipocytes white.Nat Rev Mol Cell Biol. 2016 Oct;17(10):607. doi: 10.1038/nrm.2016.125. Epub 2016 Sep 14. Nat Rev Mol Cell Biol. 2016. PMID: 27625322 No abstract available.
-
Beige Communication through Gap Junctions and Adaption by Autophagy.Cell Metab. 2016 Sep 13;24(3):370-371. doi: 10.1016/j.cmet.2016.08.024. Cell Metab. 2016. PMID: 27626195
-
Adipose tissue: Direct beige-to-white conversion.Nat Rev Endocrinol. 2016 Nov;12(11):626. doi: 10.1038/nrendo.2016.159. Epub 2016 Sep 16. Nat Rev Endocrinol. 2016. PMID: 27636727 No abstract available.
References
-
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Developmental biology. 2006;296:164–176. - PubMed
-
- Barbatelli G, Murano I, Madsen L, Hao Q, Jimenez M, Kristiansen K, Giacobino JP, De Matteis R, Cinti S. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte trans differentiation. American journal of physiology. 2010;298:E1244–1253. - PubMed
-
- Cederberg A, Gronning LM, Ahren B, Tasken K, Carlsson P, Enerback S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

