Dynamics of Nucleosome Positioning Maturation following Genomic Replication
- PMID: 27568571
- PMCID: PMC5014762
- DOI: 10.1016/j.celrep.2016.07.083
Dynamics of Nucleosome Positioning Maturation following Genomic Replication
Abstract
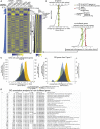
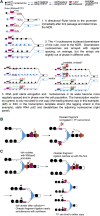
Chromatin is thought to carry epigenetic information from one generation to the next, although it is unclear how such information survives the disruptions of nucleosomal architecture occurring during genomic replication. Here, we measure a key aspect of chromatin structure dynamics during replication-how rapidly nucleosome positions are established on the newly replicated daughter genomes. By isolating newly synthesized DNA marked with 5-ethynyl-2'-deoxyuridine (EdU), we characterize nucleosome positions on both daughter genomes of S. cerevisiae during chromatin maturation. We find that nucleosomes rapidly adopt their mid-log positions at highly transcribed genes, which is consistent with a role for transcription in positioning nucleosomes in vivo. Additionally, experiments in hir1Δ mutants reveal a role for HIR in nucleosome spacing. We also characterized nucleosome positions on the leading and lagging strands, uncovering differences in chromatin maturation dynamics at hundreds of genes. Our data define the maturation dynamics of newly replicated chromatin and support a role for transcription in sculpting the chromatin template.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures






Comment in
-
Nucleosomes Find Their Place in Life.Trends Genet. 2016 Nov;32(11):689-690. doi: 10.1016/j.tig.2016.09.001. Epub 2016 Sep 17. Trends Genet. 2016. PMID: 27650123 Free PMC article.
References
-
- Alabert C., Groth A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012;13:153–167. - PubMed
-
- Alabert C., Bukowski-Wills J.C., Lee S.B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014;16:281–293. - PMC - PubMed
-
- Borodina T., Adjaye J., Sultan M. A strand-specific library preparation protocol for RNA sequencing. Methods Enzymol. 2011;500:79–98. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

