An Outbreak of Infections Caused by a Klebsiella pneumoniae ST11 Clone Coproducing Klebsiella pneumoniae Carbapenemase-2 and RmtB in a Chinese Teaching Hospital
- PMID: 27569227
- PMCID: PMC5009584
- DOI: 10.4103/0366-6999.189049
An Outbreak of Infections Caused by a Klebsiella pneumoniae ST11 Clone Coproducing Klebsiella pneumoniae Carbapenemase-2 and RmtB in a Chinese Teaching Hospital
Abstract
Background: Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae bacteria, which cause serious disease outbreaks worldwide, was rarely detected in Xiangya Hospital, prior to an outbreak that occurred from August 4, 2014, to March 17, 2015. The aim of this study was to analyze the epidemiology and molecular characteristics of the K. pneumoniae strains isolated during the outbreak.
Methods: Nonduplicate carbapenem-resistant K. pneumoniae isolates were screened for blaKPC-2and multiple other resistance determinants using polymerase chain reaction. Subsequent studies included pulsed-field gel electrophoresis (PFGE), multilocus sequence typing, analysis of plasmids, and genetic organization of blaKPC-2locus.
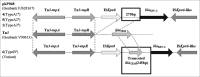
Results: Seventeen blaKPC-2-positive K. pneumoniae were identified. A wide range of resistant determinants was detected. Most isolates (88.2%) coharbored blaKPC-2and rmtB in addition to other resistance genes, including blaSHV-1, blaTEM-1, and aac(3)-IIa. The blaKPC-2and rmtB genes were located on the conjugative IncFIB-type plasmid. Genetic organization of blaKPC-2locusin most strains was consistent with that of the plasmid pKP048. Four types (A1, A2, A3, and B) were detected by PFGE, and Type A1, an ST11, was the predominant PFGE type. A novel K. pneumoniae sequence type (ST1883) related to ST11 was discovered.
Conclusions: These isolates in our study appeared to be clonal and ST11 K. pneumoniae was the predominant clone attributed to the outbreak. Coharbing of blaKPC-2and rmtB, which were located on a transferable plasmid, in clinical K. pneumoniae isolates may lead to the emergence of a new pattern of drug resistance.
Figures

Similar articles
-
A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395.Clin Microbiol Infect. 2013 Nov;19(11):E509-15. doi: 10.1111/1469-0691.12275. Epub 2013 Jul 10. Clin Microbiol Infect. 2013. PMID: 23841705
-
Clonal dissemination of multilocus sequence type 11 Klebsiella pneumoniae carbapenemase - producing K. pneumoniae in a Chinese teaching hospital.APMIS. 2015 Feb;123(2):123-7. doi: 10.1111/apm.12313. Epub 2014 Sep 25. APMIS. 2015. PMID: 25257726
-
Clonal replacement of epidemic KPC-producing Klebsiella pneumoniae in a hospital in China.BMC Infect Dis. 2017 May 23;17(1):363. doi: 10.1186/s12879-017-2467-9. BMC Infect Dis. 2017. PMID: 28535790 Free PMC article.
-
Diversity of the Genetic Environment of the blaKPC-2 Gene Among Klebsiella pneumoniae Clinical Isolates in a Chinese Hospital.Microb Drug Resist. 2016 Jan;22(1):15-21. doi: 10.1089/mdr.2014.0281. Epub 2015 Aug 27. Microb Drug Resist. 2016. PMID: 26313117
-
Carbapenemase-producing Klebsiella pneumoniae.F1000Prime Rep. 2014 Sep 4;6:80. doi: 10.12703/P6-80. eCollection 2014. F1000Prime Rep. 2014. PMID: 25343037 Free PMC article. Review.
Cited by
-
Molecular characterization and epidemiology of carbapenem non-susceptible Enterobacteriaceae isolated from the Eastern region of Heilongjiang Province, China.BMC Infect Dis. 2018 Aug 22;18(1):417. doi: 10.1186/s12879-018-3294-3. BMC Infect Dis. 2018. PMID: 30134844 Free PMC article.
-
Emergence of an NDM-5-Producing Escherichia coli Sequence Type 410 Clone in Infants in a Children's Hospital in China.Infect Drug Resist. 2020 Feb 28;13:703-710. doi: 10.2147/IDR.S244874. eCollection 2020. Infect Drug Resist. 2020. PMID: 32184632 Free PMC article.
-
Homology analysis between clinically isolated extraintestinal and enteral Klebsiella pneumoniae among neonates.BMC Microbiol. 2021 Jan 11;21(1):25. doi: 10.1186/s12866-020-02073-2. BMC Microbiol. 2021. PMID: 33430787 Free PMC article.
-
Risk Factors for Subsequential Carbapenem-Resistant Klebsiella pneumoniae Clinical Infection Among Rectal Carriers with Carbapenem-Resistant Klebsiella pneumoniae.Infect Drug Resist. 2020 May 5;13:1299-1305. doi: 10.2147/IDR.S247101. eCollection 2020. Infect Drug Resist. 2020. PMID: 32440167 Free PMC article.
-
High rate of multiresistant Klebsiella pneumoniae from human and animal origin.Infect Drug Resist. 2019 Sep 3;12:2729-2737. doi: 10.2147/IDR.S219155. eCollection 2019. Infect Drug Resist. 2019. PMID: 31564923 Free PMC article.
References
-
- Lyman M, Walters M, Lonsway D, Rasheed K, Limbago B, Kallen A. Notes from the field: Carbapenem-resistant Enterobacteriaceae producing OXA-48-like carbapenemases–United States, 2010-2015. MMWR Morb Mortal Wkly Rep. 2015;64:1315–6. doi: 10.15585/mmwr.mm6447a3. - PubMed
-
- Fursova NK, Astashkin EI, Knyazeva AI, Kartsev NN, Leonova ES, Ershova ON, et al. The spread of bla OXA-48 and bla OXA-244 carbapenemase genes among Klebsiella pneumoniae Proteus mirabilis and Enterobacter spp. isolated in Moscow, Russia. Ann Clin Microbiol Antimicrob. 2015;14:46. doi: 10.1186/s12941-015-0108-y. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

