Sex chromosome evolution in snakes inferred from divergence patterns of two gametologous genes and chromosome distribution of sex chromosome-linked repetitive sequences
- PMID: 27570632
- PMCID: PMC5002183
- DOI: 10.1186/s40851-016-0056-1
Sex chromosome evolution in snakes inferred from divergence patterns of two gametologous genes and chromosome distribution of sex chromosome-linked repetitive sequences
Abstract
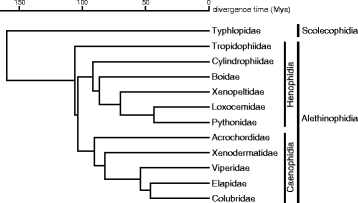
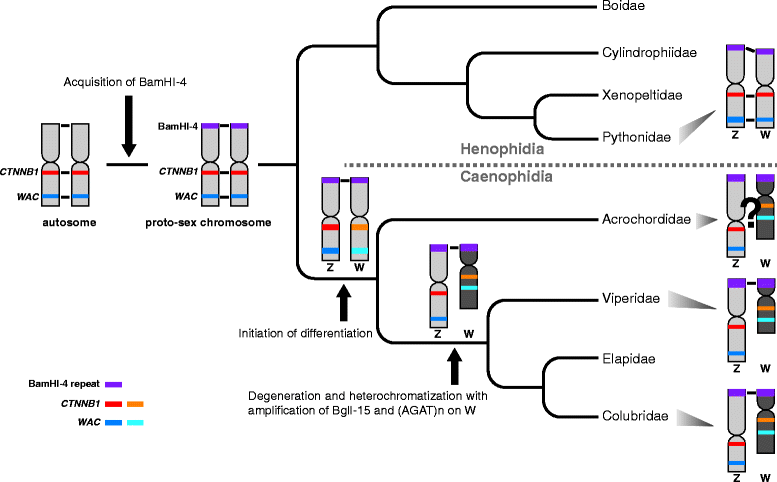
Background: The discovery of differentially organized sex chromosome systems suggests that heteromorphic sex chromosomes evolved from a pair of homologous chromosomes. Whereas karyotypes are highly conserved in alethinophidian snakes, the degeneration status of the W chromosomes varies among species. The Z and W chromosomes are morphologically homomorphic in henophidian species, whereas in snakes belonging to caenophidian families the W chromosomes are highly degenerated. Snakes therefore are excellent animal models in which to study sex chromosome evolution. Herein, we investigated the differentiation processes for snake sex chromosomes using both coding and repetitive sequences. We analyzed phylogenetic relationships of CTNNB1 and WAC genes, localized to the centromeric and telomeric regions, respectively, of the long arms on snake sex chromosomes, and chromosome distribution of sex chromosome-linked repetitive sequences in several henophidian and caenophidian species.
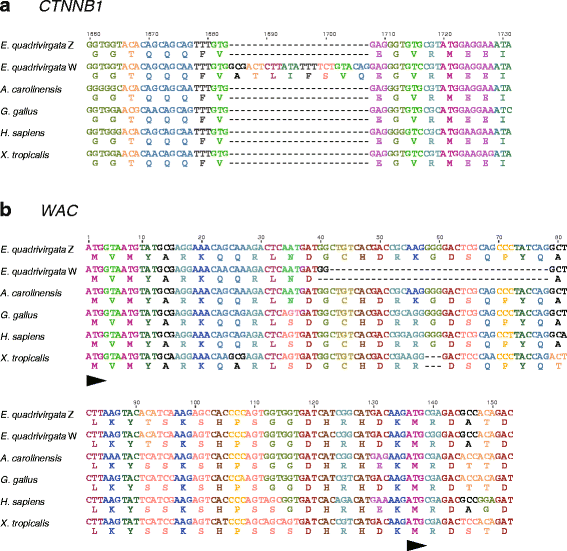
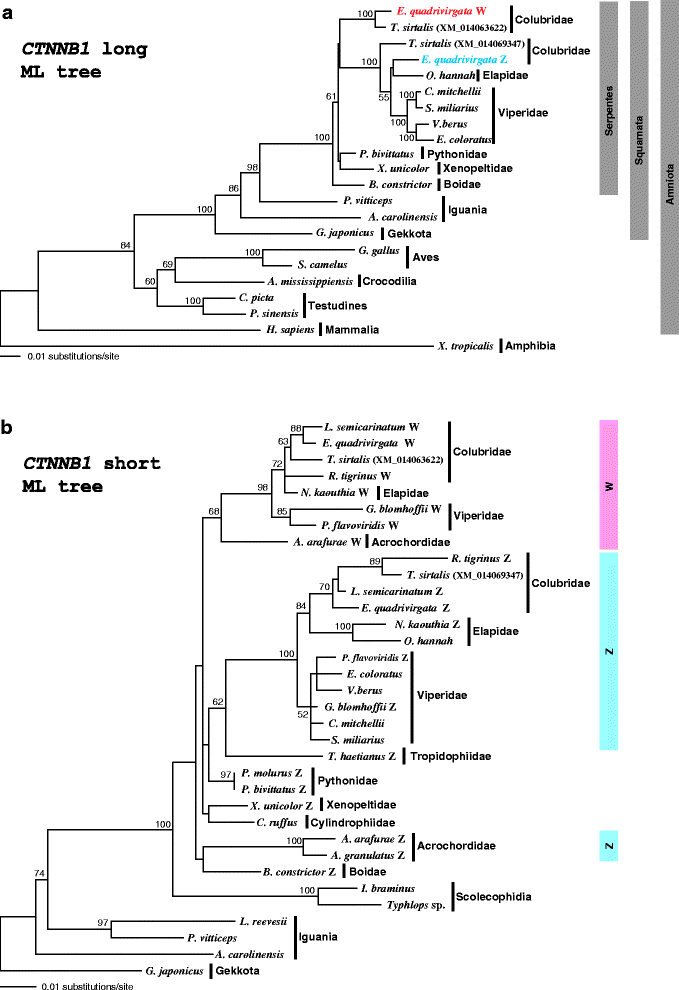
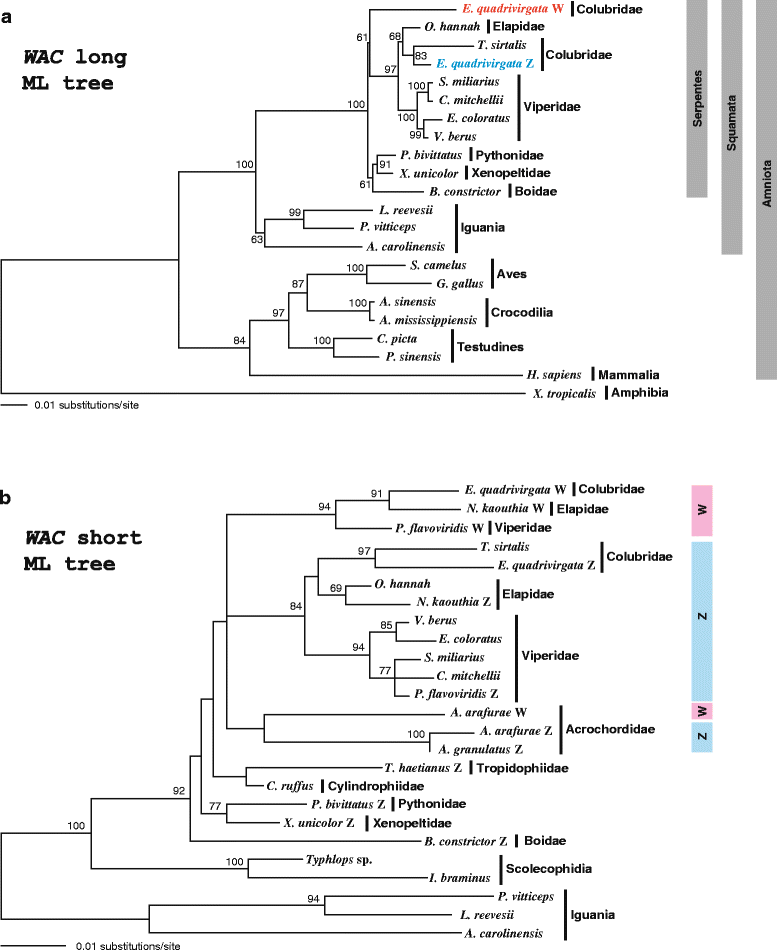
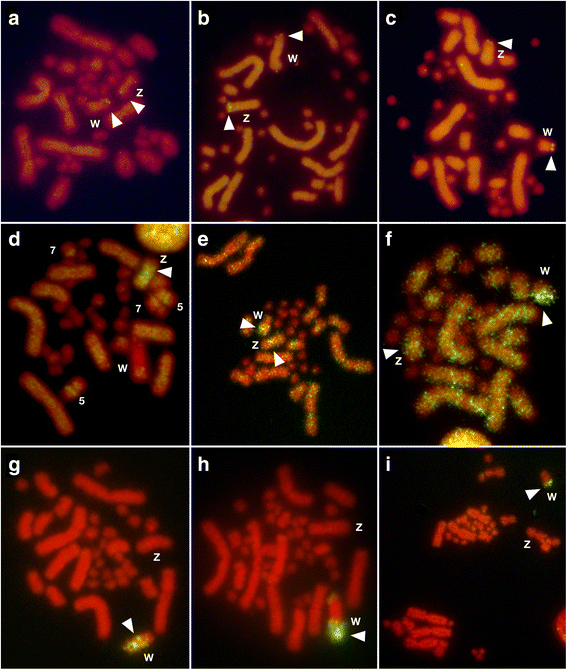
Results: Partial or full-length coding sequences of CTNNB1 and WAC were identified for Z homologs of henophidian species from Tropidophiidae, Boidae, Cylindrophiidae, Xenopeltidae, and Pythonidae, and for Z and W homologs of caenophidian species from Acrochordidae, Viperidae, Elapidae, and Colubridae. Female-specific sequences for the two genes were not found in the henophidian (boid and pythonid) species examined. Phylogenetic trees constructed using each gene showed that the Z and W homologs of the caenophidian species cluster separately. The repetitive sequence isolated from the W chromosome heterochromatin of the colubrid Elaphe quadrivirgata and a microsatellite motif (AGAT)8 were strongly hybridized with W chromosomes of the viperid and colubrid species examined.
Conclusion: Our phylogenetic analyses suggest that the cessation of recombination between the Z and W homologs of CTNNB1 and WAC predated the diversification of the caenophidian families. As the repetitive sequences on the W chromosomes were shared among viperid and colubrid species, heterochromatinization of the proto-W chromosome appears to have occurred before the splitting of these two groups. These results collectively suggest that differentiation of the proto-Z and proto-W chromosomes extended to wide regions on the sex chromosomes in the common ancestor of caenophidian families during a relatively short period.
Keywords: Evolution; Gametolog; Heterochromatin; Phylogeny; Repetitive sequences; Snake; W chromosome; Z chromosome.
Figures
References
-
- Muller HJ. A gene for the fourth chromosome of Drosophila. J Exp Zool. 1914;17:325–336. doi: 10.1002/jez.1400170303. - DOI
-
- Ohno S. Sex chromosomes and sex-linked genes. Berlin: Springer; 1967.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

