Mutation of the Enterohemorrhagic Escherichia coli Core LPS Biosynthesis Enzyme RfaD Confers Hypersusceptibility to Host Intestinal Innate Immunity In vivo
- PMID: 27570746
- PMCID: PMC4982379
- DOI: 10.3389/fcimb.2016.00082
Mutation of the Enterohemorrhagic Escherichia coli Core LPS Biosynthesis Enzyme RfaD Confers Hypersusceptibility to Host Intestinal Innate Immunity In vivo
Abstract
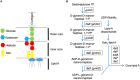
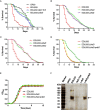
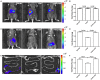
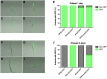
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is an important foodborne pathogen causing severe diseases in humans worldwide. Currently, there is no specific treatment available for EHEC infection and the use of conventional antibiotics is contraindicated. Therefore, identification of potential therapeutic targets and development of effective measures to control and treat EHEC infection are needed. Lipopolysaccharides (LPS) are surface glycolipids found on the outer membrane of gram-negative bacteria, including EHEC, and LPS biosynthesis has long been considered as potential anti-bacterial target. Here, we demonstrated that the EHEC rfaD gene that functions in the biosynthesis of the LPS inner core is required for the intestinal colonization and pathogenesis of EHEC in vivo. Disruption of the EHEC rfaD confers attenuated toxicity in Caenorhabditis elegans and less bacterial colonization in the intestine of C. elegans and mouse. Moreover, rfaD is also involved in the control of susceptibility of EHEC to antimicrobial peptides and host intestinal immunity. It is worth noting that rfaD mutation did not interfere with the growth kinetics when compared to the wild-type EHEC cells. Taken together, we demonstrated that mutations of the EHEC rfaD confer hypersusceptibility to host intestinal innate immunity in vivo, and suggested that targeting the RfaD or the core LPS synthesis pathway may provide alternative therapeutic regimens for EHEC infection.
Keywords: Caenorhabditis elegans; RfaD/GmhD/WaaD; antimicrobial peptides (AMPs); enterohemorrhagic Escherichia coli (EHEC); intestinal innate immunity; lipopolysaccharide (LPS).
Figures



References
-
- Armstrong G. D., Mulvey G. L., Marcato P., Griener T. P., Kahan M. C., Tennent G. A., et al. . (2006). Human Serum Amyloid P component protects against Escherichia coli O157:H7 Shiga Toxin 2 in vivo: therapeutic implications for hemolytic-uremic syndrome. J. Infect. Dis. 193, 1120–1124. 10.1086/501472 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

