Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder
- PMID: 27571009
- PMCID: PMC5841760
- DOI: 10.1038/nn.4373
Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder
Abstract
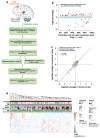
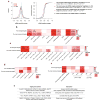
Genetic variants conferring risk for autism spectrum disorder (ASD) have been identified, but the role of post-transcriptional mechanisms in ASD is not well understood. We performed genome-wide microRNA (miRNA) expression profiling in post-mortem brains from individuals with ASD and controls and identified miRNAs and co-regulated modules that were perturbed in ASD. Putative targets of these ASD-affected miRNAs were enriched for genes that have been implicated in ASD risk. We confirmed regulatory relationships between several miRNAs and their putative target mRNAs in primary human neural progenitors. These include hsa-miR-21-3p, a miRNA of unknown CNS function that is upregulated in ASD and that targets neuronal genes downregulated in ASD, and hsa_can_1002-m, a previously unknown, primate-specific miRNA that is downregulated in ASD and that regulates the epidermal growth factor receptor and fibroblast growth factor receptor signaling pathways involved in neural development and immune function. Our findings support a role for miRNA dysregulation in ASD pathophysiology and provide a rich data set and framework for future analyses of miRNAs in neuropsychiatric diseases.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

