Cognate HLA absence in trans diminishes human NK cell education
- PMID: 27571408
- PMCID: PMC5096830
- DOI: 10.1172/JCI86923
Cognate HLA absence in trans diminishes human NK cell education
Abstract
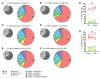
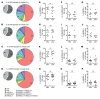
NK cells are innate lymphocytes with protective functions against viral infections and tumor formation. Human NK cells carry inhibitory killer cell Ig-like receptors (KIRs), which recognize distinct HLAs. NK cells with KIRs for self-HLA molecules acquire superior cytotoxicity against HLA- tumor cells during education for improved missing-self recognition. Here, we reconstituted mice with human hematopoietic cells from donors with homozygous KIR ligands or with a mix of hematopoietic cells from these homozygous donors, allowing assessment of the resulting KIR repertoire and NK cell education. We found that co-reconstitution with 2 KIR ligand-mismatched compartments did not alter the frequency of KIR-expressing NK cells. However, NK cell education was diminished in mice reconstituted with parallel HLA compartments due to a lack of cognate HLA molecules on leukocytes for the corresponding KIRs. This change in NK cell education in mixed human donor-reconstituted mice improved NK cell-mediated immune control of EBV infection, indicating that mixed hematopoietic cell populations could be exploited to improve NK cell reactivity against leukotropic pathogens. Taken together, these findings indicate that leukocytes lacking cognate HLA ligands can disarm KIR+ NK cells in a manner that may decrease HLA- tumor cell recognition but allows for improved NK cell-mediated immune control of a human γ-herpesvirus.
Figures





References
-
- Trinchieri G, Santoli D. Anti-viral activity induced by culturing lymphocytes with tumor-derived or virus-transformed cells. Enhancement of human natural killer cell activity by interferon and antagonistic inhibition of susceptibility of target cells to lysis. J Exp Med. 1978;147(5):1314–1333. doi: 10.1084/jem.147.5.1314. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

