Crystal structure of Clostridium difficile toxin A
- PMID: 27571750
- PMCID: PMC4976693
- DOI: 10.1038/nmicrobiol.2015.2
Crystal structure of Clostridium difficile toxin A
Abstract
Clostridium difficile infection is the leading cause of hospital-acquired diarrhoea and pseudomembranous colitis. Disease is mediated by the actions of two toxins, TcdA and TcdB, which cause the diarrhoea, as well as inflammation and necrosis within the colon. The toxins are large (308 and 270 kDa, respectively), homologous (47% amino acid identity) glucosyltransferases that target small GTPases within the host. The multidomain toxins enter cells by receptor-mediated endocytosis and, upon exposure to the low pH of the endosome, insert into and deliver two enzymatic domains across the membrane. Eukaryotic inositol-hexakisphosphate (InsP6) binds an autoprocessing domain to activate a proteolysis event that releases the N-terminal glucosyltransferase domain into the cytosol. Here, we report the crystal structure of a 1,832-amino-acid fragment of TcdA (TcdA1832), which reveals a requirement for zinc in the mechanism of toxin autoprocessing and an extended delivery domain that serves as a scaffold for the hydrophobic α-helices involved in pH-dependent pore formation. A surface loop of the delivery domain whose sequence is strictly conserved among all large clostridial toxins is shown to be functionally important, and is highlighted for future efforts in the development of vaccines and novel therapeutics.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
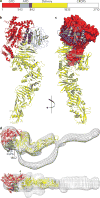


Similar articles
-
Crystal structure of Clostridium difficile toxin A.Nat Microbiol. 2016;1:15002. doi: 10.1038/nmicrobiol.2015.2. Epub 2016 Jan 11. Nat Microbiol. 2016. PMID: 27512603
-
The antimicrobial peptide Angie 5 inhibits TcdA and TcdB from Clostridioides difficile.Cell Mol Life Sci. 2025 Jun 30;82(1):265. doi: 10.1007/s00018-025-05799-2. Cell Mol Life Sci. 2025. PMID: 40586877 Free PMC article.
-
Characterization of Flagellum and Toxin Phase Variation in Clostridioides difficile Ribotype 012 Isolates.J Bacteriol. 2018 Jun 25;200(14):e00056-18. doi: 10.1128/JB.00056-18. Print 2018 Jul 15. J Bacteriol. 2018. PMID: 29735765 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Antibiotic treatment for Clostridium difficile-associated diarrhoea in adults.Cochrane Database Syst Rev. 2017 Mar 3;3(3):CD004610. doi: 10.1002/14651858.CD004610.pub5. Cochrane Database Syst Rev. 2017. PMID: 28257555 Free PMC article.
Cited by
-
Identification of TFPI as a receptor reveals recombination-driven receptor switching in Clostridioides difficile toxin B variants.Nat Commun. 2022 Nov 9;13(1):6786. doi: 10.1038/s41467-022-33964-9. Nat Commun. 2022. PMID: 36351897 Free PMC article.
-
Novel structural insights for a pair of monoclonal antibodies recognizing non-overlapping epitopes of the glucosyltransferase domain of Clostridium difficile toxin B.Curr Res Struct Biol. 2022 Apr 7;4:96-105. doi: 10.1016/j.crstbi.2022.03.003. eCollection 2022. Curr Res Struct Biol. 2022. PMID: 35469152 Free PMC article.
-
Clostridium difficile ribotype 017 - characterization, evolution and epidemiology of the dominant strain in Asia.Emerg Microbes Infect. 2019;8(1):796-807. doi: 10.1080/22221751.2019.1621670. Emerg Microbes Infect. 2019. PMID: 31138041 Free PMC article. Review.
-
A neutralizing antibody that blocks delivery of the enzymatic cargo of Clostridium difficile toxin TcdB into host cells.J Biol Chem. 2018 Jan 19;293(3):941-952. doi: 10.1074/jbc.M117.813428. Epub 2017 Nov 27. J Biol Chem. 2018. PMID: 29180448 Free PMC article.
-
Infection: Modulation of Clostridium difficile infection by dietary zinc.Nat Rev Gastroenterol Hepatol. 2016 Dec;13(12):686-688. doi: 10.1038/nrgastro.2016.177. Epub 2016 Nov 9. Nat Rev Gastroenterol Hepatol. 2016. PMID: 27826138 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

