Oncogenic roles of the SETDB2 histone methyltransferase in gastric cancer
- PMID: 27572307
- PMCID: PMC5341872
- DOI: 10.18632/oncotarget.11625
Oncogenic roles of the SETDB2 histone methyltransferase in gastric cancer
Abstract
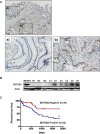
SETDB2 is a histone H3 lysine 9 (H3K9) tri-methyltransferase that is involved in transcriptional gene silencing. Since it is still unknown whether SETDB2 is linked to carcinogenesis, we studied alterations and functions of SETDB2 in human gastric cancers (GCs). SETDB2 protein was highly expressed in 30 of 72 (41.7%) primary GC tissues compared with their normal counterparts by immunohistochemistry. SETDB2 overexpression was significantly associated with the late stage of GCs (P<0.05) and poor prognosis of GC patients (P<0.05). The GC cell lines with SETDB2 knockdown and overexpression significantly decreased and increased cell proliferation, migration and invasion, respectively (P<0.05). Knockdown of SETDB2 in MKN74 and MKN45 cells reduced global H3K9 tri-methylation (me3) levels. Microarray analysis indicated that expression of WWOX and CADM1, tumor suppressor genes, was significantly enhanced in MKN74 cells after SETDB2 knockdown. Chromatin immunoprecipitation assays showed that the H3K9me3 levels at the promoter regions of these two genes corresponded to the SETDB2 expression levels in GC cells. Moreover, ectopic SETDB2 protein was recruited to their promoter regions. Our data suggest that SETDB2 is associated with transcriptional repression of WWOX and CADM1, and hence overexpression of SETDB2 may contribute to GC progression.
Keywords: H3K9me3; SETDB2; gastric cancer; histone methyltransferase.
Conflict of interest statement
All authors declare that they have no conflict of interest to disclose.
Figures





References
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. - PubMed
-
- Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat Rev Cancer. 2003;3:592–600. Review. - PubMed
-
- Wanajo A, Sasaki A, Nagasaki H, Shimada S, Otsubo T, Owaki S, Shimizu Y, Eishi Y, Kojima K, Nakajima Y, Kawano T, Yuasa Y, Akiyama Y. Methylation of the calcium channel-related gene, CACNA2D3, is frequent and a poor prognostic factor in gastric cancer. Gastroenterology. 2008;35:580–590. - PubMed
-
- Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21:209–220. - PubMed
-
- Rodriguez-Paredes M, Martinez de Paz A, Simó-Riudalbas L, Sayols S, Moutinho C, Moran S, Villanueva A, Vázquez-Cedeira M, Lazo PA, Carneiro F, Moura CS, Vieira J, Teixeira MR, et al. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene. 2014;33:2807–2813. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

