Metabolomic Profiling of the Malaria Box Reveals Antimalarial Target Pathways
- PMID: 27572391
- PMCID: PMC5075069
- DOI: 10.1128/AAC.01224-16
Metabolomic Profiling of the Malaria Box Reveals Antimalarial Target Pathways
Abstract
The threat of widespread drug resistance to frontline antimalarials has renewed the urgency for identifying inexpensive chemotherapeutic compounds that are effective against Plasmodium falciparum, the parasite species responsible for the greatest number of malaria-related deaths worldwide. To aid in the fight against malaria, a recent extensive screening campaign has generated thousands of lead compounds with low micromolar activity against blood stage parasites. A subset of these leads has been compiled by the Medicines for Malaria Venture (MMV) into a collection of structurally diverse compounds known as the MMV Malaria Box. Currently, little is known regarding the activity of these Malaria Box compounds on parasite metabolism during intraerythrocytic development, and a majority of the targets for these drugs have yet to be defined. Here we interrogated the in vitro metabolic effects of 189 drugs (including 169 of the drug-like compounds from the Malaria Box) using ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS). The resulting metabolic fingerprints provide information on the parasite biochemical pathways affected by pharmacologic intervention and offer a critical blueprint for selecting and advancing lead compounds as next-generation antimalarial drugs. Our results reveal several major classes of metabolic disruption, which allow us to predict the mode of action (MoA) for many of the Malaria Box compounds. We anticipate that future combination therapies will be greatly informed by these results, allowing for the selection of appropriate drug combinations that simultaneously target multiple metabolic pathways, with the aim of eliminating malaria and forestalling the expansion of drug-resistant parasites in the field.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


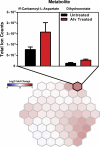
References
-
- RTS,S Clinical Trials Partnership. 2015. Efficacy and safety of RTS,S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet 386:31–45. doi:10.1016/S0140-6736(15)60721-8. - DOI - PMC - PubMed
-
- Gamo FJ, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera JL, Vanderwall DE, Green DV, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. 2010. Thousands of chemical starting points for antimalarial lead identification. Nature 465:305–310. doi:10.1038/nature09107. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

