Molecular Epidemiology of Mutations in Antimicrobial Resistance Loci of Pseudomonas aeruginosa Isolates from Airways of Cystic Fibrosis Patients
- PMID: 27572404
- PMCID: PMC5075079
- DOI: 10.1128/AAC.00724-16
Molecular Epidemiology of Mutations in Antimicrobial Resistance Loci of Pseudomonas aeruginosa Isolates from Airways of Cystic Fibrosis Patients
Abstract
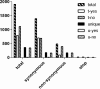
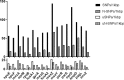
The chronic airway infections with Pseudomonas aeruginosa in people with cystic fibrosis (CF) are treated with aerosolized antibiotics, oral fluoroquinolones, and/or intravenous combination therapy with aminoglycosides and β-lactam antibiotics. An international strain collection of 361 P. aeruginosa isolates from 258 CF patients seen at 30 CF clinics was examined for mutations in 17 antimicrobial susceptibility and resistance loci that had been identified as hot spots of mutation by genome sequencing of serial isolates from a single CF clinic. Combinatorial amplicon sequencing of pooled PCR products identified 1,112 sequence variants that were not present in the genomes of representative strains of the 20 most common clones of the global P. aeruginosa population. A high frequency of singular coding variants was seen in spuE, mexA, gyrA, rpoB, fusA1, mexZ, mexY, oprD, ampD, parR, parS, and envZ (amgS), reflecting the pressure upon P. aeruginosa in lungs of CF patients to generate novel protein variants. The proportion of nonneutral amino acid exchanges was high. Of the 17 loci, mexA, mexZ, and pagL were most frequently affected by independent stop mutations. Private and de novo mutations seem to play a pivotal role in the response of P. aeruginosa populations to the antimicrobial load and the individual CF host.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Mogayzel PJ Jr., Naureckas ET, Robinson KA, Brady C, Guill M, Lahiri T, Lubsch L, Matsui J, Oermann CM, Ratjen F, Rosenfeld M, Simon RH, Hazle L, Sabadosa K, Marshall BC; Cystic Fibrosis Foundation Pulmonary Clinical Practice Guidelines Committee. 2014. Cystic Fibrosis Foundation pulmonary guideline. Pharmacologic approaches to prevention and eradication of initial Pseudomonas aeruginosa infection. Ann Am Thorac Soc 11:1640–1650. doi:10.1513/AnnalsATS.201404-166OC. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

