Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome
- PMID: 27572443
- PMCID: PMC5031140
- DOI: 10.1038/nmicrobiol.2016.24
Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome
Abstract
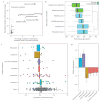
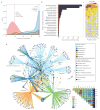
Development of the preterm infant gut microbiota is emerging as a critical research priority(1). Since preterm infants almost universally receive early and often extended antibiotic therapy(2), it is important to understand how these interventions alter gut microbiota development(3-6). Analysis of 401 stools from 84 longitudinally sampled preterm infants demonstrates that meropenem, cefotaxime and ticarcillin-clavulanate are associated with significantly reduced species richness. In contrast, vancomycin and gentamicin, the antibiotics most commonly administered to preterm infants, have non-uniform effects on species richness, but these can be predicted with 85% accuracy based on the relative abundance of only two bacterial species and two antibiotic resistance (AR) genes at treatment initiation. To investigate resistome development, we functionally selected resistance to 16 antibiotics from 21 faecal metagenomic expression libraries. Of the 794 AR genes identified, 79% had not previously been classified as AR genes. Combined with deep shotgun sequencing of all stools, we find that multidrug-resistant members of the genera Escherichia, Klebsiella and Enterobacter, genera commonly associated with nosocomial infections, dominate the preterm infant gut microbiota. AR genes that are enriched following specific antibiotic treatments are generally unique to the specific treatment and are highly correlated with the abundance of a single species. The most notable exceptions include ticarcillin-clavulanate and ampicillin, both of which enrich for a large number of overlapping AR genes, and are correlated with Klebsiella pneumoniae. We find that all antibiotic treatments are associated with widespread collateral microbiome impact by enrichment of AR genes that have no known activity against the specific antibiotic driver.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
Antibiotics, gut bugs and the young.Nat Rev Microbiol. 2016 Jun;14(6):336. doi: 10.1038/nrmicro.2016.73. Epub 2016 May 3. Nat Rev Microbiol. 2016. PMID: 27140687
-
Microbiome: Antibiotics and the infant microflora.Nat Microbiol. 2016 Mar 29;1:16040. doi: 10.1038/nmicrobiol.2016.40. Nat Microbiol. 2016. PMID: 27572452 No abstract available.
References
-
- Clark RH, Bloom BT, Spitzer AR, Gerstmann DR. Reported medication use in the neonatal intensive care unit: data from a large national data set. Pediatrics. 2006;117:1979–1987. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

