Influenza virus mRNA trafficking through host nuclear speckles
- PMID: 27572970
- PMCID: PMC4917225
- DOI: 10.1038/nmicrobiol.2016.69
Influenza virus mRNA trafficking through host nuclear speckles
Erratum in
-
Corrigendum: Influenza virus mRNA trafficking through host nuclear speckles.Nat Microbiol. 2017 Jan 9;2:17002. doi: 10.1038/nmicrobiol.2017.2. Nat Microbiol. 2017. PMID: 28067239 No abstract available.
Abstract
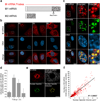
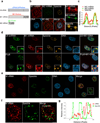
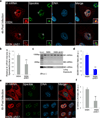
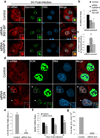
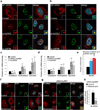
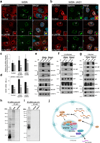
Influenza A virus is a human pathogen with a genome composed of eight viral RNA segments that replicate in the nucleus. Two viral mRNAs are alternatively spliced. The unspliced M1 mRNA is translated into the matrix M1 protein, while the ion channel M2 protein is generated after alternative splicing. These proteins are critical mediators of viral trafficking and budding. We show that the influenza virus uses nuclear speckles to promote post-transcriptional splicing of its M1 mRNA. We assign previously unknown roles for the viral NS1 protein and cellular factors to an intranuclear trafficking pathway that targets the viral M1 mRNA to nuclear speckles, mediates splicing at these nuclear bodies and exports the spliced M2 mRNA from the nucleus. Given that nuclear speckles are storage sites for splicing factors, which leave these sites to splice cellular pre-mRNAs at transcribing genes, we reveal a functional subversion of nuclear speckles to promote viral gene expression.
Keywords: influenza virus; mRMA export; nuclear speckles; splicing.
Figures
Comment in
-
Viral cell biology: Influenza raids the splicing store.Nat Microbiol. 2016 Jun 24;1(7):16100. doi: 10.1038/nmicrobiol.2016.100. Nat Microbiol. 2016. PMID: 27572980 No abstract available.
References
-
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

