Requirements for Pseudomonas aeruginosa Type I-F CRISPR-Cas Adaptation Determined Using a Biofilm Enrichment Assay
- PMID: 27573013
- PMCID: PMC5075037
- DOI: 10.1128/JB.00458-16
Requirements for Pseudomonas aeruginosa Type I-F CRISPR-Cas Adaptation Determined Using a Biofilm Enrichment Assay
Abstract
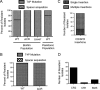
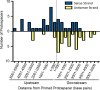
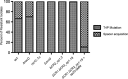
CRISPR (clustered regularly interspaced short palindromic repeat)-Cas (CRISPR-associated protein) systems are diverse and found in many archaea and bacteria. These systems have mainly been characterized as adaptive immune systems able to protect against invading mobile genetic elements, including viruses. The first step in this protection is acquisition of spacer sequences from the invader DNA and incorporation of those sequences into the CRISPR array, termed CRISPR adaptation. Progress in understanding the mechanisms and requirements of CRISPR adaptation has largely been accomplished using overexpression of cas genes or plasmid loss assays; little work has focused on endogenous CRISPR-acquired immunity from viral predation. Here, we developed a new biofilm-based assay system to enrich for Pseudomonas aeruginosa strains with new spacer acquisition. We used this assay to demonstrate that P. aeruginosa rapidly acquires spacers protective against DMS3vir, an engineered lytic variant of the Mu-like bacteriophage DMS3, through primed CRISPR adaptation from spacers present in the native CRISPR2 array. We found that for the P. aeruginosa type I-F system, the cas1 gene is required for CRISPR adaptation, recG contributes to (but is not required for) primed CRISPR adaptation, recD is dispensable for primed CRISPR adaptation, and finally, the ability of a putative priming spacer to prime can vary considerably depending on the specific sequences of the spacer.
Importance: Our understanding of CRISPR adaptation has expanded largely through experiments in type I CRISPR systems using plasmid loss assays, mutants of Escherichia coli, or cas1-cas2 overexpression systems, but there has been little focus on studying the adaptation of endogenous systems protecting against a lytic bacteriophage. Here we describe a biofilm system that allows P. aeruginosa to rapidly gain spacers protective against a lytic bacteriophage. This approach has allowed us to probe the requirements for CRISPR adaptation in the endogenous type I-F system of P. aeruginosa Our data suggest that CRISPR-acquired immunity in a biofilm may be one reason that many P. aeruginosa strains maintain a CRISPR-Cas system.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Cas1 and Cas2 From the Type II-C CRISPR-Cas System of Riemerella anatipestifer Are Required for Spacer Acquisition.Front Cell Infect Microbiol. 2018 Jun 12;8:195. doi: 10.3389/fcimb.2018.00195. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29951376 Free PMC article.
-
Clustered Regularly Interspaced Short Palindromic Repeat-Dependent, Biofilm-Specific Death of Pseudomonas aeruginosa Mediated by Increased Expression of Phage-Related Genes.mBio. 2015 May 12;6(3):e00129-15. doi: 10.1128/mBio.00129-15. mBio. 2015. PMID: 25968642 Free PMC article.
-
Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery.Nucleic Acids Res. 2015 Dec 15;43(22):10848-60. doi: 10.1093/nar/gkv1261. Epub 2015 Nov 19. Nucleic Acids Res. 2015. PMID: 26586803 Free PMC article.
-
Analysis of direct repeats and spacers of CRISPR/Cas systems type I-F in Brazilian clinical strains of Pseudomonas aeruginosa.Mol Genet Genomics. 2019 Oct;294(5):1095-1105. doi: 10.1007/s00438-019-01575-7. Epub 2019 May 16. Mol Genet Genomics. 2019. PMID: 31098740 Review.
-
CRISPR-Cas adaptation in Escherichia coli.Biosci Rep. 2023 Mar 31;43(3):BSR20221198. doi: 10.1042/BSR20221198. Biosci Rep. 2023. PMID: 36809461 Free PMC article. Review.
Cited by
-
Comparative Analysis of CRISPR-Cas Systems in Pseudomonas Genomes.Genes (Basel). 2023 Jun 25;14(7):1337. doi: 10.3390/genes14071337. Genes (Basel). 2023. PMID: 37510242 Free PMC article.
-
Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity.Proc Natl Acad Sci U S A. 2017 Jun 27;114(26):E5113-E5121. doi: 10.1073/pnas.1616395114. Epub 2017 Apr 24. Proc Natl Acad Sci U S A. 2017. PMID: 28438998 Free PMC article.
-
Complete Genome Sequence of Pseudomonas aeruginosa K34-7, a Carbapenem-Resistant Isolate of the High-Risk Sequence Type 233.Microbiol Resour Announc. 2018 Aug 2;7(4):e00886-18. doi: 10.1128/MRA.00886-18. eCollection 2018 Aug. Microbiol Resour Announc. 2018. PMID: 30533874 Free PMC article.
-
Genomic features of "Candidatus Venteria ishoeyi", a new sulfur-oxidizing macrobacterium from the Humboldt Sulfuretum off Chile.PLoS One. 2017 Dec 13;12(12):e0188371. doi: 10.1371/journal.pone.0188371. eCollection 2017. PLoS One. 2017. PMID: 29236755 Free PMC article.
-
Phage susceptibility to a minimal, modular synthetic CRISPR-Cas system in Pseudomonas aeruginosa is nutrient dependent.Philos Trans R Soc Lond B Biol Sci. 2025 Sep 4;380(1934):20240473. doi: 10.1098/rstb.2024.0473. Epub 2025 Sep 4. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 40904105 Free PMC article.
References
-
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736. doi:10.1038/nrmicro3569. - DOI - PMC - PubMed
-
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. 2015. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell 60:385–397. doi:10.1016/j.molcel.2015.10.008. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

