Lys-63-linked Ubiquitination of γ-Aminobutyric Acid (GABA), Type B1, at Multiple Sites by the E3 Ligase Mind Bomb-2 Targets GABAB Receptors to Lysosomal Degradation
- PMID: 27573246
- PMCID: PMC5076837
- DOI: 10.1074/jbc.M116.750968
Lys-63-linked Ubiquitination of γ-Aminobutyric Acid (GABA), Type B1, at Multiple Sites by the E3 Ligase Mind Bomb-2 Targets GABAB Receptors to Lysosomal Degradation
Abstract
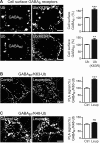
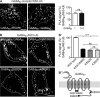
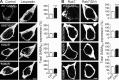
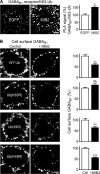
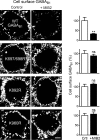
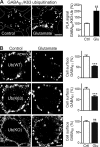
GABAB receptors are heterodimeric G protein-coupled receptors, which control neuronal excitability by mediating prolonged inhibition. The magnitude of GABAB receptor-mediated inhibition essentially depends on the amount of receptors in the plasma membrane. However, the factors regulating cell surface expression of GABAB receptors are poorly characterized. Cell surface GABAB receptors are constitutively internalized and either recycled to the plasma membrane or degraded in lysosomes. The signal that sorts GABAB receptors to lysosomes is currently unknown. Here we show that Mind bomb-2 (MIB2)-mediated Lys-63-linked ubiquitination of the GABAB1 subunit at multiple sites is the lysosomal sorting signal for GABAB receptors. We found that inhibition of lysosomal activity in cultured rat cortical neurons increased the fraction of Lys-63-linked ubiquitinated GABAB receptors and enhanced the expression of total as well as cell surface GABAB receptors. Mutational inactivation of four putative ubiquitination sites in the GABAB1 subunit significantly diminished Lys-63-linked ubiquitination of GABAB receptors and prevented their lysosomal degradation. We identified MIB2 as the E3 ligase triggering Lys-63-linked ubiquitination and lysosomal degradation of GABAB receptors. Finally, we show that sustained activation of glutamate receptors, a condition occurring in brain ischemia that down-regulates GABAB receptors, considerably increased the expression of MIB2 and Lys-63-linked ubiquitination of GABAB receptors. Interfering with Lys-63-linked ubiquitination by overexpressing ubiquitin mutants or GABAB1 mutants deficient in Lys-63-linked ubiquitination prevented glutamate-induced down-regulation of the receptors. These findings indicate that Lys-63-linked ubiquitination of GABAB1 at multiple sites by MIB2 controls sorting of GABAB receptors to lysosomes for degradation under physiological and pathological conditions.
Keywords: E3 ubiquitin ligase; GABA receptor; lysosome; protein degradation; ubiquitylation (ubiquitination).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





Similar articles
-
Ca2+/Calmodulin-Dependent Protein Kinase II (CaMKII) β-Dependent Phosphorylation of GABAB1 Triggers Lysosomal Degradation of GABAB Receptors via Mind Bomb-2 (MIB2)-Mediated Lys-63-Linked Ubiquitination.Mol Neurobiol. 2019 Feb;56(2):1293-1309. doi: 10.1007/s12035-018-1142-5. Epub 2018 Jun 7. Mol Neurobiol. 2019. PMID: 29881949 Free PMC article.
-
Endoplasmic reticulum-associated degradation controls cell surface expression of γ-aminobutyric acid, type B receptors.J Biol Chem. 2013 Nov 29;288(48):34897-905. doi: 10.1074/jbc.M113.514745. Epub 2013 Oct 10. J Biol Chem. 2013. PMID: 24114844 Free PMC article.
-
Post-endocytotic Deubiquitination and Degradation of the Metabotropic γ-Aminobutyric Acid Receptor by the Ubiquitin-specific Protease 14.J Biol Chem. 2016 Mar 25;291(13):7156-70. doi: 10.1074/jbc.M115.686907. Epub 2016 Jan 27. J Biol Chem. 2016. PMID: 26817839 Free PMC article.
-
Endo-lysosomal sorting of G-protein-coupled receptors by ubiquitin: Diverse pathways for G-protein-coupled receptor destruction and beyond.Traffic. 2019 Feb;20(2):101-109. doi: 10.1111/tra.12619. Epub 2018 Nov 18. Traffic. 2019. PMID: 30353650 Free PMC article. Review.
-
Hepatic cytochromes P450: structural degrons and barcodes, posttranslational modifications and cellular adapters in the ERAD-endgame.Drug Metab Rev. 2016 Aug;48(3):405-33. doi: 10.1080/03602532.2016.1195403. Epub 2016 Jun 20. Drug Metab Rev. 2016. PMID: 27320797 Free PMC article. Review.
Cited by
-
Role of Rab10 in cocaine-induced behavioral effects is associated with GABAB receptor membrane expression in the nucleus accumbens.Front Pharmacol. 2024 Nov 27;15:1496657. doi: 10.3389/fphar.2024.1496657. eCollection 2024. Front Pharmacol. 2024. PMID: 39669198 Free PMC article.
-
Diversity of structure and function of GABAB receptors: a complexity of GABAB-mediated signaling.Proc Jpn Acad Ser B Phys Biol Sci. 2018;94(10):390-411. doi: 10.2183/pjab.94.026. Proc Jpn Acad Ser B Phys Biol Sci. 2018. PMID: 30541966 Free PMC article. Review.
-
ERK1/2-Dependent Phosphorylation of GABAB1(S867/T872), Controlled by CaMKIIβ, Is Required for GABAB Receptor Degradation under Physiological and Pathological Conditions.Int J Mol Sci. 2023 Aug 30;24(17):13436. doi: 10.3390/ijms241713436. Int J Mol Sci. 2023. PMID: 37686242 Free PMC article.
-
E3 Ubiquitin Ligase RNF125 Activates Interleukin-36 Receptor Signaling and Contributes to Its Turnover.J Innate Immun. 2018;10(1):56-69. doi: 10.1159/000481210. Epub 2017 Nov 25. J Innate Immun. 2018. PMID: 29176319 Free PMC article.
-
Protein Modifications with Ubiquitin as Response to Cerebral Ischemia-Reperfusion Injury.Transl Stroke Res. 2018 Apr;9(2):157-173. doi: 10.1007/s12975-017-0567-x. Epub 2017 Aug 25. Transl Stroke Res. 2018. PMID: 28842824 Review.
References
-
- Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 - PubMed
-
- Pinard A., Seddik R., and Bettler B. (2010) GABAB receptors: physiological functions and mechanisms of diversity. Adv. Pharmacol. 58, 231–255 - PubMed
-
- Kohl M. M., and Paulsen O. (2010) The roles of GABAB receptors in cortical network activity. Adv. Pharmacol. 58, 205–229 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

