The epigenome in Alzheimer's disease: current state and approaches for a new path to gene discovery and understanding disease mechanism
- PMID: 27573688
- PMCID: PMC5189639
- DOI: 10.1007/s00401-016-1612-7
The epigenome in Alzheimer's disease: current state and approaches for a new path to gene discovery and understanding disease mechanism
Abstract
The advent of new technologies and analytic approaches is beginning to provide an unprecedented look at features of the human genome that affect RNA expression. These "epigenomic" features are found in a number of different forms: they include DNA methylation, covalent modifications of histone proteins and non-coding RNAs. Some of these features have now been implicated in Alzheimer's disease (AD). Here, we focus on recent studies that have identified robust observations relating to DNA methylation and chromatin in human brain tissue; these findings will ground the next generation of studies and provide a model for the design of such studies. Stemming from observations that compounds with histone deacetylase activity may be beneficial in AD, epigenome-wide studies in cortical samples from large numbers of human subjects have now shown that AD-associated epigenomic changes are reproducible, are not driven by genetic risk factors, and are widespread at specific locations in the genome. A fundamental question of whether such changes are causal remains to be demonstrated, but it is already clear that well-powered investigations of the human epigenome in the target organ of a neurodegenerative disease are feasible, are implicating new areas of the genome in the disease, and will be an important tool for future studies. We are now at an inflection point: as genome-wide association studies of genetic variants come to an end, a new generation of studies exploring the epigenome will provide an important new layer of information with which to enrich our understanding of AD pathogenesis and to possibly guide development of new therapeutic targets.
Keywords: Aging; Alzheimer’s disease; Chromatin; DNA methylation; Epigenomics; Histone.
Figures
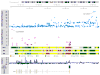
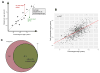
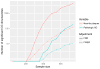
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

