Healthy human gut phageome
- PMID: 27573828
- PMCID: PMC5027468
- DOI: 10.1073/pnas.1601060113
Healthy human gut phageome
Abstract
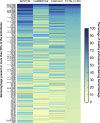
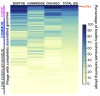
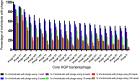
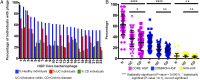
The role of bacteriophages in influencing the structure and function of the healthy human gut microbiome is unknown. With few exceptions, previous studies have found a high level of heterogeneity in bacteriophages from healthy individuals. To better estimate and identify the shared phageome of humans, we analyzed a deep DNA sequence dataset of active bacteriophages and available metagenomic datasets of the gut bacteriophage community from healthy individuals. We found 23 shared bacteriophages in more than one-half of 64 healthy individuals from around the world. These shared bacteriophages were found in a significantly smaller percentage of individuals with gastrointestinal/irritable bowel disease. A network analysis identified 44 bacteriophage groups of which 9 (20%) were shared in more than one-half of all 64 individuals. These results provide strong evidence of a healthy gut phageome (HGP) in humans. The bacteriophage community in the human gut is a mixture of three classes: a set of core bacteriophages shared among more than one-half of all people, a common set of bacteriophages found in 20-50% of individuals, and a set of bacteriophages that are either rarely shared or unique to a person. We propose that the core and common bacteriophage communities are globally distributed and comprise the HGP, which plays an important role in maintaining gut microbiome structure/function and thereby contributes significantly to human health.
Keywords: gut microbiome bacteriophage; gut microbiome viruses; human gut viral metagenome; shared microbiome viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

