Anatomy of the β-branching enzyme of polyketide biosynthesis and its interaction with an acyl-ACP substrate
- PMID: 27573844
- PMCID: PMC5027445
- DOI: 10.1073/pnas.1607210113
Anatomy of the β-branching enzyme of polyketide biosynthesis and its interaction with an acyl-ACP substrate
Abstract
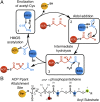
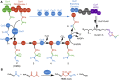
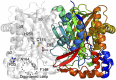
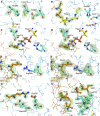
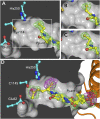
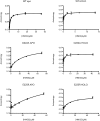
Alkyl branching at the β position of a polyketide intermediate is an important variation on canonical polyketide natural product biosynthesis. The branching enzyme, 3-hydroxy-3-methylglutaryl synthase (HMGS), catalyzes the aldol addition of an acyl donor to a β-keto-polyketide intermediate acceptor. HMGS is highly selective for two specialized acyl carrier proteins (ACPs) that deliver the donor and acceptor substrates. The HMGS from the curacin A biosynthetic pathway (CurD) was examined to establish the basis for ACP selectivity. The donor ACP (CurB) had high affinity for the enzyme (Kd = 0.5 μM) and could not be substituted by the acceptor ACP. High-resolution crystal structures of HMGS alone and in complex with its donor ACP reveal a tight interaction that depends on exquisite surface shape and charge complementarity between the proteins. Selectivity is explained by HMGS binding to an unusual surface cleft on the donor ACP, in a manner that would exclude the acceptor ACP. Within the active site, HMGS discriminates between pre- and postreaction states of the donor ACP. The free phosphopantetheine (Ppant) cofactor of ACP occupies a conserved pocket that excludes the acetyl-Ppant substrate. In comparison with HMG-CoA (CoA) synthase, the homologous enzyme from primary metabolism, HMGS has several differences at the active site entrance, including a flexible-loop insertion, which may account for the specificity of one enzyme for substrates delivered by ACP and the other by CoA.
Keywords: HMG synthase; acyl carrier protein; curacin; natural products; polyketide synthase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Weissman KJ. Genetic engineering of modular PKSs: From combinatorial biosynthesis to synthetic biology. Nat Prod Rep. 2016;33(2):203–230. - PubMed
-
- Calderone CT. Isoprenoid-like alkylations in polyketide biosynthesis. Nat Prod Rep. 2008;25(5):845–853. - PubMed
-
- Blokhin AV, et al. Characterization of the interaction of the marine cyanobacterial natural product curacin A with the colchicine site of tubulin and initial structure-activity studies with analogues. Mol Pharmacol. 1995;48(3):523–531. - PubMed
-
- Verdier-Pinard P, et al. Structure-activity analysis of the interaction of curacin A, the potent colchicine site antimitotic agent, with tubulin and effects of analogs on the growth of MCF-7 breast cancer cells. Mol Pharmacol. 1998;53(1):62–76. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

