Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon
- PMID: 27573849
- PMCID: PMC5027439
- DOI: 10.1073/pnas.1607327113
Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon
Abstract
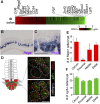
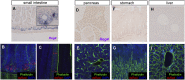
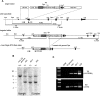
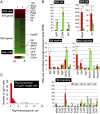
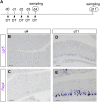
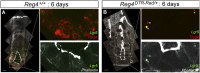
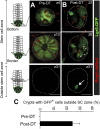
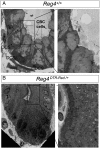
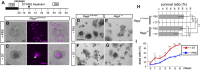
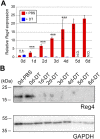
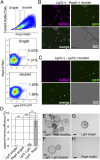
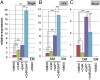
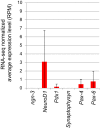
Leucine-rich repeat-containing G-protein coupled receptor 5-positive (Lgr5(+)) stem cells reside at crypt bottoms of the small and large intestine. Small intestinal Paneth cells supply Wnt3, EGF, and Notch signals to neighboring Lgr5(+) stem cells. Whereas the colon lacks Paneth cells, deep crypt secretory (DCS) cells are intermingled with Lgr5(+) stem cells at crypt bottoms. Here, we report regenerating islet-derived family member 4 (Reg4) as a marker of DCS cells. To investigate a niche function, we eliminated DCS cells by using the diphtheria-toxin receptor gene knocked into the murine Reg4 locus. Ablation of DCS cells results in loss of stem cells from colonic crypts and disrupts gut homeostasis and colon organoid growth. In agreement, sorted Reg4(+) DCS cells promote organoid formation of single Lgr5(+) colon stem cells. DCS cells can be massively produced from Lgr5(+) colon stem cells in vitro by combined Notch inhibition and Wnt activation. We conclude that Reg4(+) DCS cells serve as Paneth cell equivalents in the colon crypt niche.
Keywords: Lgr5; Reg4; deep crypt secretory cells; intestinal stem cell; niche.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

