Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing
- PMID: 27573852
- PMCID: PMC5027458
- DOI: 10.1073/pnas.1522203113
Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing
Abstract
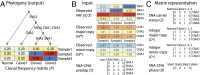
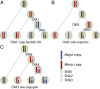
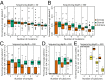
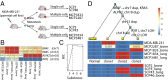
Cancer is a disease driven by evolutionary selection on somatic genetic and epigenetic alterations. Here, we propose Canopy, a method for inferring the evolutionary phylogeny of a tumor using both somatic copy number alterations and single-nucleotide alterations from one or more samples derived from a single patient. Canopy is applied to bulk sequencing datasets of both longitudinal and spatial experimental designs and to a transplantable metastasis model derived from human cancer cell line MDA-MB-231. Canopy successfully identifies cell populations and infers phylogenies that are in concordance with existing knowledge and ground truth. Through simulations, we explore the effects of key parameters on deconvolution accuracy and compare against existing methods. Canopy is an open-source R package available at https://cran.r-project.org/web/packages/Canopy/.
Keywords: cancer evolution; cancer genomics; clonal deconvolution; intratumor heterogeneity; phylogeny inference.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Multiregional Tumor Trees Are Not Phylogenies.Trends Cancer. 2017 Aug;3(8):546-550. doi: 10.1016/j.trecan.2017.06.004. Epub 2017 Jul 17. Trends Cancer. 2017. PMID: 28780931 Free PMC article.
-
Inferring clonal evolution of tumors from single nucleotide somatic mutations.BMC Bioinformatics. 2014 Feb 1;15:35. doi: 10.1186/1471-2105-15-35. BMC Bioinformatics. 2014. PMID: 24484323 Free PMC article.
-
Integrative inference of subclonal tumour evolution from single-cell and bulk sequencing data.Nat Commun. 2019 Jun 21;10(1):2750. doi: 10.1038/s41467-019-10737-5. Nat Commun. 2019. PMID: 31227714 Free PMC article.
-
Principles of Reconstructing the Subclonal Architecture of Cancers.Cold Spring Harb Perspect Med. 2017 Aug 1;7(8):a026625. doi: 10.1101/cshperspect.a026625. Cold Spring Harb Perspect Med. 2017. PMID: 28270531 Free PMC article. Review.
-
Computational Approaches for the Investigation of Intra-tumor Heterogeneity and Clonal Evolution from Bulk Sequencing Data in Precision Oncology Applications.Adv Exp Med Biol. 2022;1361:101-118. doi: 10.1007/978-3-030-91836-1_6. Adv Exp Med Biol. 2022. PMID: 35230685 Review.
Cited by
-
Engineering Heterogeneous Tumor Models for Biomedical Applications.Adv Sci (Weinh). 2024 Jan;11(1):e2304160. doi: 10.1002/advs.202304160. Epub 2023 Nov 9. Adv Sci (Weinh). 2024. PMID: 37946674 Free PMC article. Review.
-
Implications of non-uniqueness in phylogenetic deconvolution of bulk DNA samples of tumors.Algorithms Mol Biol. 2019 Sep 3;14:19. doi: 10.1186/s13015-019-0155-6. eCollection 2019. Algorithms Mol Biol. 2019. PMID: 31497065 Free PMC article.
-
Exploring Current Challenges and Perspectives for Automatic Reconstruction of Clonal Evolution.Cancer Genomics Proteomics. 2022 Mar-Apr;19(2):194-204. doi: 10.21873/cgp.20314. Cancer Genomics Proteomics. 2022. PMID: 35181588 Free PMC article.
-
Cumulative smoking dose affects the clinical outcomes of EGFR-mutated lung adenocarcinoma patients treated with EGFR-TKIs: a retrospective study.BMC Cancer. 2018 Jul 28;18(1):768. doi: 10.1186/s12885-018-4691-0. BMC Cancer. 2018. PMID: 30055587 Free PMC article.
-
Estimation of cancer cell fractions and clone trees from multi-region sequencing of tumors.Bioinformatics. 2022 Aug 2;38(15):3677-3683. doi: 10.1093/bioinformatics/btac367. Bioinformatics. 2022. PMID: 35642899 Free PMC article.
References
-
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. - PubMed
-
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9(4):138–141. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

