Degeneration of aflatoxin gene clusters in Aspergillus flavus from Africa and North America
- PMID: 27576895
- PMCID: PMC5005231
- DOI: 10.1186/s13568-016-0228-6
Degeneration of aflatoxin gene clusters in Aspergillus flavus from Africa and North America
Abstract
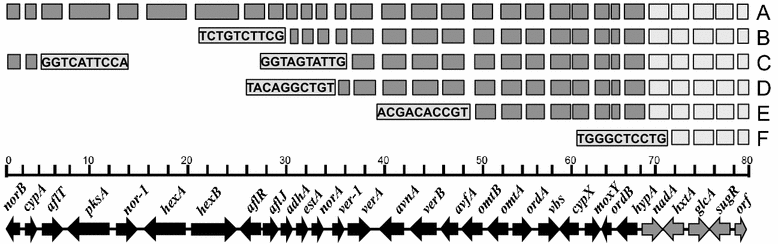
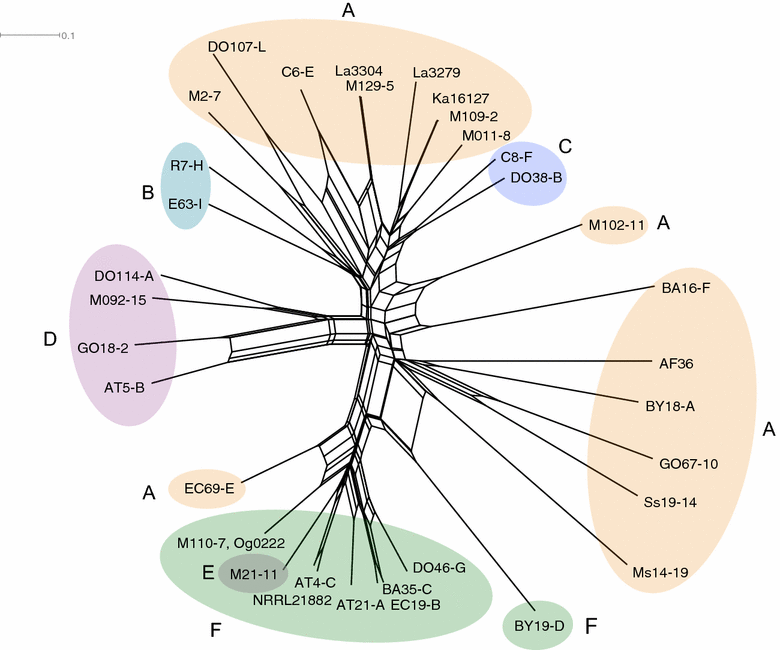
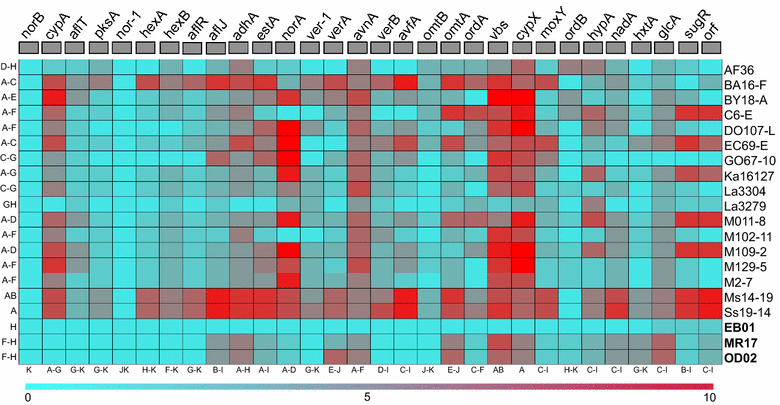
Aspergillus flavus is the most common causal agent of aflatoxin contamination of food and feed. However, aflatoxin-producing potential varies widely among A. flavus genotypes with many producing no aflatoxins. Some non-aflatoxigenic genotypes are used as biocontrol agents to prevent contamination. Aflatoxin biosynthesis genes are tightly clustered in a highly conserved order. Gene deletions and presence of single nucleotide polymorphisms (SNPs) in aflatoxin biosynthesis genes are often associated with A. flavus inability to produce aflatoxins. In order to identify mechanisms of non-aflatoxigenicity in non-aflatoxigenic genotypes of value in aflatoxin biocontrol, complete cluster sequences of 35 A. flavus genotypes from Africa and North America were analyzed. Inability of some genotypes to produce aflatoxin resulted from deletion of biosynthesis genes. In other genotypes, non-aflatoxigenicity originated from SNP formation. The process of degeneration differed across the gene cluster; genes involved in early biosynthesis stages were more likely to be deleted while genes involved in later stages displayed high frequencies of SNPs. Comparative analyses of aflatoxin gene clusters provides insight into the diversity of mechanisms of non-aflatoxigenicity in A. flavus genotypes used as biological control agents. The sequences provide resources for both diagnosis of non-aflatoxigenicity and monitoring of biocontrol genotypes during biopesticide manufacture and in the environment.
Keywords: Aflatoxin gene cluster; Aspergillus flavus; Biocontrol; Cluster degeneration; Evolution; Non-aflatoxigenic.
Figures

References
-
- Antilla L, Cotty PJ. The ARS-ACRPC partnership to control aflatoxin in Arizona cotton: current status. Mycopathologia. 2002;155:64.
-
- Atehnkeng J, Ojiambo PS, Cotty PJ, Bandyopadhyay R. Field efficacy of a mixture of atoxigenic Aspergillus flavus Link: Fr vegetative compatibility groups in preventing aflatoxin contamination in maize (Zea mays L.) Biol Control. 2014;72:62–70. doi: 10.1016/j.biocontrol.2014.02.009. - DOI
-
- Bandyopadhyay R, Cotty PJ. Biological controls for aflatoxin reduction, vol 2020 Vision Focus 20(16). Washington DC: International Food Policty Research Institute (IFPRI); 2013.
LinkOut - more resources
Full Text Sources
Other Literature Sources

