Hypoxia inducible factor-1 mediates the expression of the immune checkpoint HLA-G in glioma cells through hypoxia response element located in exon 2
- PMID: 27577073
- PMCID: PMC5325396
- DOI: 10.18632/oncotarget.11628
Hypoxia inducible factor-1 mediates the expression of the immune checkpoint HLA-G in glioma cells through hypoxia response element located in exon 2
Abstract
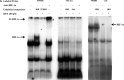
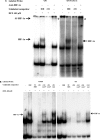
HLA-G is an immune checkpoint molecule with specific relevance in cancer immunotherapy. It was first identified in cytotrophoblasts, protecting the fetus from maternal rejection. HLA-G tissue expression is very restricted but induced in numerous malignant tumors such as glioblastoma, contributing to their immune escape. Hypoxia occurs during placenta and tumor development and was shown to activate HLA-G. We aimed to elucidate the mechanisms of HLA-G activation under conditions combining hypoxia-mimicking treatment and 5-aza-2'deoxycytidine, a DNA demethylating agent used in anti-cancer therapy which also induces HLA-G. Both treatments enhanced the amount of HLA-G mRNA and protein in HLA-G negative U251MG glioma cells. Electrophoretic Mobility Shift Assays and luciferase reporter gene assays revealed that HLA-G upregulation depends on Hypoxia Inducible Factor-1 (HIF-1) and a hypoxia responsive element (HRE) located in exon 2. A polymorphic HRE at -966 bp in the 5'UT region may modulate the magnitude of the response mediated by the exon 2 HRE. We suggest that therapeutic strategies should take into account that HLA-G expression in response to hypoxic tumor environment is dependent on HLA-G gene polymorphism and DNA methylation state at the HLA-G locus.
Keywords: HIF-1; HLA-G; exon 2 HRE; glioma.
Conflict of interest statement
None.
Figures






Similar articles
-
Structural Characterization of the Interaction of Hypoxia Inducible Factor-1 with Its Hypoxia Responsive Element at the -964G > A Variation Site of the HLA-G Promoter Region.Int J Mol Sci. 2021 Dec 2;22(23):13046. doi: 10.3390/ijms222313046. Int J Mol Sci. 2021. PMID: 34884849 Free PMC article.
-
Human leukocyte antigen-G is frequently expressed in glioblastoma and may be induced in vitro by combined 5-aza-2'-deoxycytidine and interferon-γ treatments: results from a multicentric study.Am J Pathol. 2013 Feb;182(2):540-52. doi: 10.1016/j.ajpath.2012.10.021. Epub 2012 Dec 4. Am J Pathol. 2013. PMID: 23219427 Free PMC article.
-
Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway.Cancer Res. 2002 Aug 1;62(15):4316-24. Cancer Res. 2002. PMID: 12154035
-
Hypoxic Modulation of HLA-G Expression through the Metabolic Sensor HIF-1 in Human Cancer Cells.J Immunol Res. 2017;2017:4587520. doi: 10.1155/2017/4587520. Epub 2017 Jul 11. J Immunol Res. 2017. PMID: 28781970 Free PMC article. Review.
-
Computational and atomistic studies applied to the understanding of the structural and behavioral features of the immune checkpoint HLA-G molecule and gene.Hum Immunol. 2023 Aug;84(8):374-383. doi: 10.1016/j.humimm.2023.01.004. Epub 2023 Jan 27. Hum Immunol. 2023. PMID: 36710086 Review.
Cited by
-
Mechanisms of HIF-driven immunosuppression in tumour microenvironment.J Egypt Natl Canc Inst. 2023 Aug 30;35(1):27. doi: 10.1186/s43046-023-00186-z. J Egypt Natl Canc Inst. 2023. PMID: 37646847 Review.
-
HIF: a master regulator of nutrient availability and metabolic cross-talk in the tumor microenvironment.EMBO J. 2023 Mar 15;42(6):e112067. doi: 10.15252/embj.2022112067. Epub 2023 Feb 20. EMBO J. 2023. PMID: 36808622 Free PMC article. Review.
-
Hypoxia-Driven Immunosuppressive Metabolites in the Tumor Microenvironment: New Approaches for Combinational Immunotherapy.Front Immunol. 2018 Jul 16;9:1591. doi: 10.3389/fimmu.2018.01591. eCollection 2018. Front Immunol. 2018. PMID: 30061885 Free PMC article. Review.
-
Differentially Expressed Bone Marrow microRNAs Are Associated With Soluble HLA-G Bone Marrow Levels in Childhood Leukemia.Front Genet. 2022 Jun 14;13:871972. doi: 10.3389/fgene.2022.871972. eCollection 2022. Front Genet. 2022. PMID: 35774498 Free PMC article.
-
Integration of Epigenetic Mechanisms into Non-Genotoxic Carcinogenicity Hazard Assessment: Focus on DNA Methylation and Histone Modifications.Int J Mol Sci. 2021 Oct 11;22(20):10969. doi: 10.3390/ijms222010969. Int J Mol Sci. 2021. PMID: 34681626 Free PMC article. Review.
References
-
- Mouillot G, Marcou C, Zidi I, Guillard C, Sangrouber D, Carosella ED, Moreau P. Hypoxia modulates HLA-G gene expression in tumor cells. Hum Immunol. 2007;68:277–285. - PubMed
-
- Chang CC, Murphy SP, Ferrone S. Differential in vivo and in vitro HLA-G expression in melanoma cells: potential mechanisms. Hum Immunol. 2003;64:1057–1063. - PubMed
-
- Patel R, Shervington L, Lea R, Shervington A. Epigenetic silencing of telomerase and a non-alkylating agent as a novel therapeutic approach for glioma. Brain Res. 2008;1188:173–181. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

