Performance comparison of two commercial human whole-exome capture systems on formalin-fixed paraffin-embedded lung adenocarcinoma samples
- PMID: 27578032
- PMCID: PMC5004269
- DOI: 10.1186/s12885-016-2720-4
Performance comparison of two commercial human whole-exome capture systems on formalin-fixed paraffin-embedded lung adenocarcinoma samples
Abstract
Background: Next Generation Sequencing (NGS) has become a valuable tool for molecular landscape characterization of cancer genomes, leading to a better understanding of tumor onset and progression, and opening new avenues in translational oncology. Formalin-fixed paraffin-embedded (FFPE) tissue is the method of choice for storage of clinical samples, however low quality of FFPE genomic DNA (gDNA) can limit its use for downstream applications.
Methods: To investigate the FFPE specimen suitability for NGS analysis and to establish the performance of two solution-based exome capture technologies, we compared the whole-exome sequencing (WES) data of gDNA extracted from 5 fresh frozen (FF) and 5 matched FFPE lung adenocarcinoma tissues using: SeqCap EZ Human Exome v.3.0 (Roche NimbleGen) and SureSelect XT Human All Exon v.5 (Agilent Technologies).
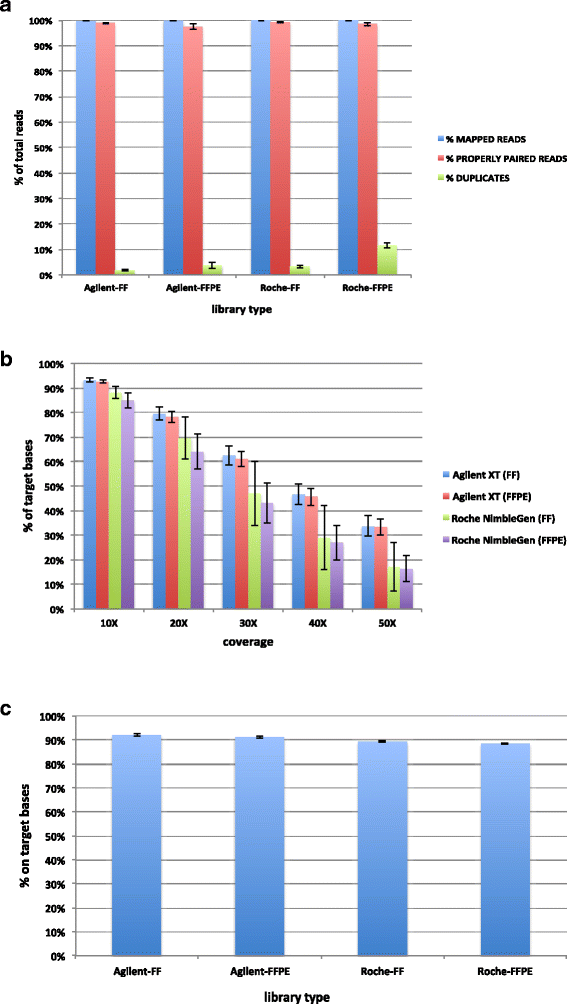
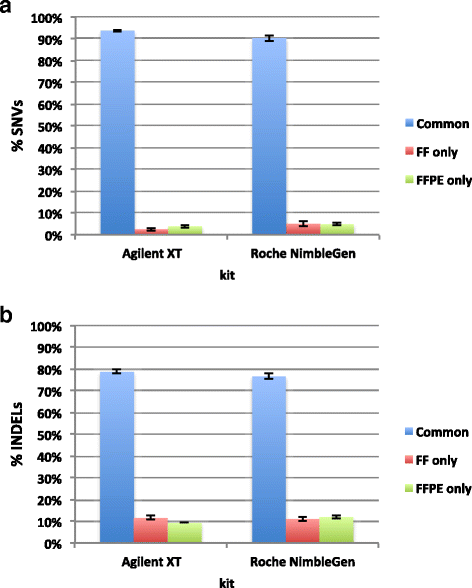
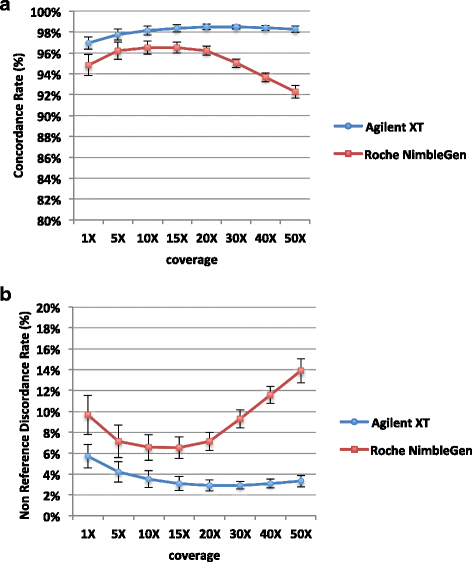
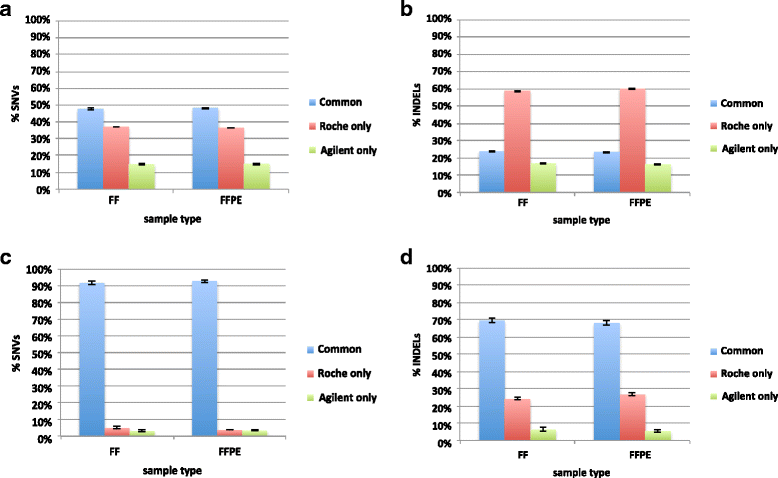
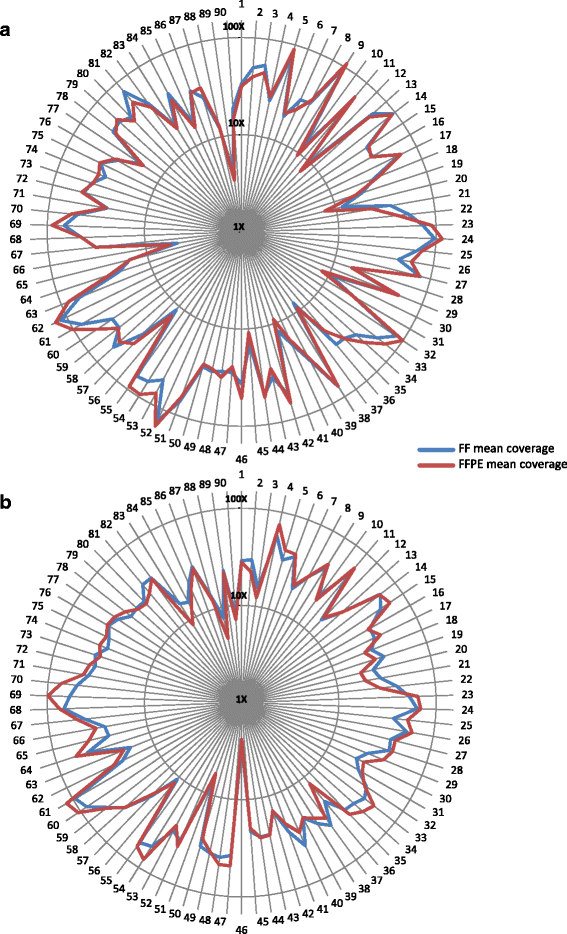
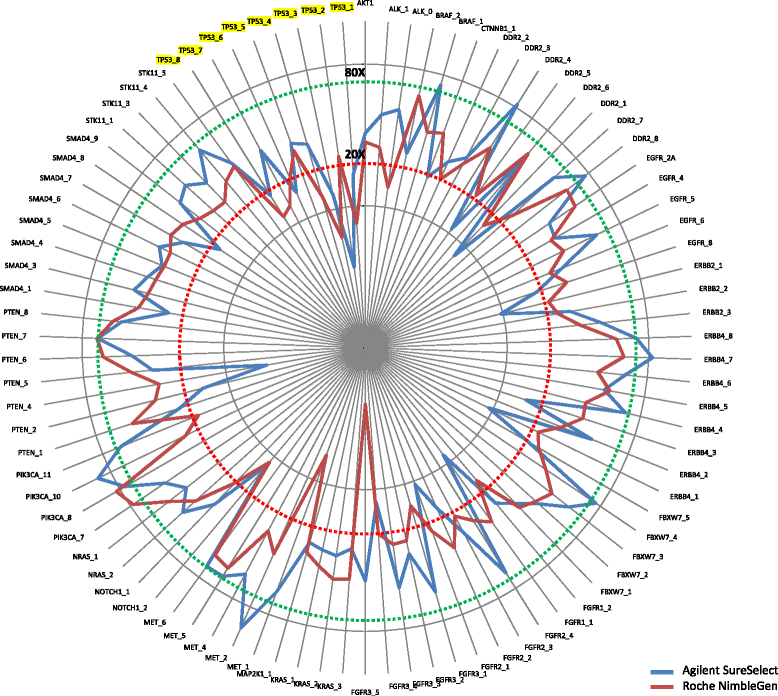
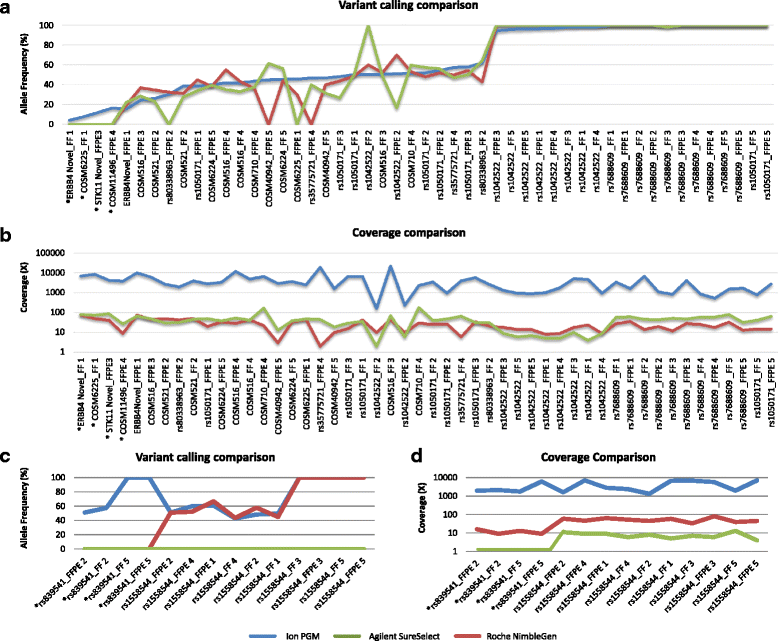
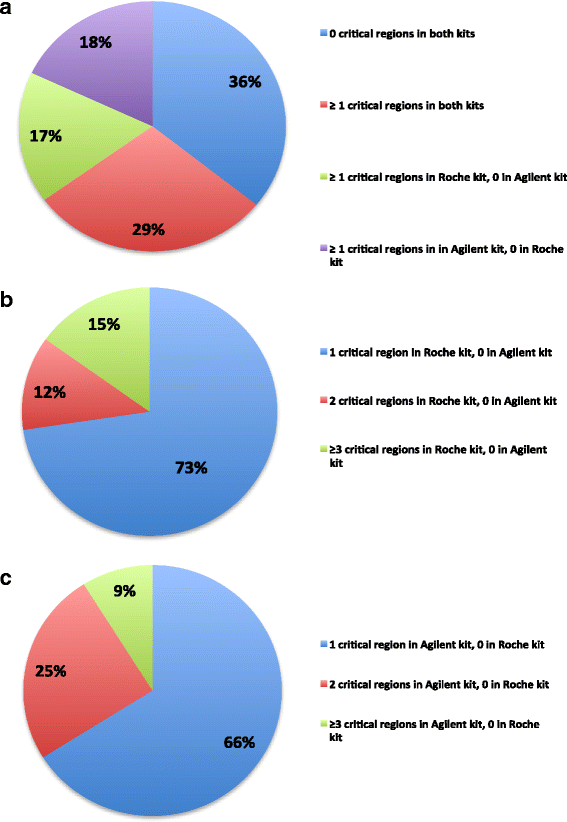
Results: Sequencing metrics on Illumina HiSeq were optimal for both exome systems and comparable among FFPE and FF samples, with a slight increase of PCR duplicates in FFPE, mainly in Roche NimbleGen libraries. Comparison of single nucleotide variants (SNVs) between FFPE-FF pairs reached overlapping values >90 % in both systems. Both WES showed high concordance with target re-sequencing data by Ion PGM™ in 22 lung-cancer genes, regardless the source of samples. Exon coverage of 623 cancer-related genes revealed high coverage efficiency of both kits, proposing WES as a valid alternative to target re-sequencing.
Conclusions: High-quality and reliable data can be successfully obtained from WES of FFPE samples starting from a relatively low amount of input gDNA, suggesting the inclusion of NGS-based tests into clinical contest. In conclusion, our analysis suggests that the WES approach could be extended to a translational research context as well as to the clinic (e.g. to study rare malignancies), where the simultaneous analysis of the whole coding region of the genome may help in the detection of cancer-linked variants.
Keywords: Cancer-related genes; Exome sequencing; FFPE; Lung adenocarcinoma; Quality control; Solution-based capture.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

