A High-Coverage Yersinia pestis Genome from a Sixth-Century Justinianic Plague Victim
- PMID: 27578768
- PMCID: PMC5062324
- DOI: 10.1093/molbev/msw170
A High-Coverage Yersinia pestis Genome from a Sixth-Century Justinianic Plague Victim
Abstract
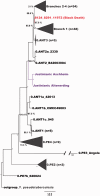
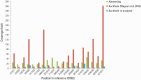
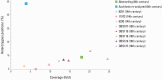
The Justinianic Plague, which started in the sixth century and lasted to the mid eighth century, is thought to be the first of three historically documented plague pandemics causing massive casualties. Historical accounts and molecular data suggest the bacterium Yersinia pestis as its etiological agent. Here we present a new high-coverage (17.9-fold) Y. pestis genome obtained from a sixth-century skeleton recovered from a southern German burial site close to Munich. The reconstructed genome enabled the detection of 30 unique substitutions as well as structural differences that have not been previously described. We report indels affecting a lacl family transcription regulator gene as well as nonsynonymous substitutions in the nrdE, fadJ, and pcp genes, that have been suggested as plague virulence determinants or have been shown to be upregulated in different models of plague infection. In addition, we identify 19 false positive substitutions in a previously published lower-coverage Y. pestis genome from another archaeological site of the same time period and geographical region that is otherwise genetically identical to the high-coverage genome sequence reported here, suggesting low-genetic diversity of the plague during the sixth century in rural southern Germany.
Keywords: Yersinia pestis; ancient DNA; justinianic plague; reconstruction; whole genome.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

Comment in
-
Reconstructing the Sixth Century Plague from a Victim.Mol Biol Evol. 2016 Nov;33(11):3028-3029. doi: 10.1093/molbev/msw203. Epub 2016 Oct 10. Mol Biol Evol. 2016. PMID: 27738270 No abstract available.
Similar articles
-
Ancient Yersinia pestis genomes from across Western Europe reveal early diversification during the First Pandemic (541-750).Proc Natl Acad Sci U S A. 2019 Jun 18;116(25):12363-12372. doi: 10.1073/pnas.1820447116. Epub 2019 Jun 4. Proc Natl Acad Sci U S A. 2019. PMID: 31164419 Free PMC article.
-
Yersinia pestis DNA from skeletal remains from the 6(th) century AD reveals insights into Justinianic Plague.PLoS Pathog. 2013;9(5):e1003349. doi: 10.1371/journal.ppat.1003349. Epub 2013 May 2. PLoS Pathog. 2013. PMID: 23658525 Free PMC article.
-
Yersinia pestis strains from Latvia show depletion of the pla virulence gene at the end of the second plague pandemic.Sci Rep. 2020 Sep 3;10(1):14628. doi: 10.1038/s41598-020-71530-9. Sci Rep. 2020. PMID: 32884081 Free PMC article.
-
Historical plague pandemics: perspectives from ancient DNA.Trends Microbiol. 2025 Jan;33(1):7-10. doi: 10.1016/j.tim.2024.10.008. Epub 2024 Nov 29. Trends Microbiol. 2025. PMID: 39613690 Review.
-
Genome and Evolution of Yersinia pestis.Adv Exp Med Biol. 2016;918:171-192. doi: 10.1007/978-94-024-0890-4_6. Adv Exp Med Biol. 2016. PMID: 27722863 Review.
Cited by
-
Yersinia pestis: the Natural History of Plague.Clin Microbiol Rev. 2020 Dec 9;34(1):e00044-19. doi: 10.1128/CMR.00044-19. Print 2020 Dec 16. Clin Microbiol Rev. 2020. PMID: 33298527 Free PMC article. Review.
-
A genomic and historical synthesis of plague in 18th century Eurasia.Proc Natl Acad Sci U S A. 2020 Nov 10;117(45):28328-28335. doi: 10.1073/pnas.2009677117. Epub 2020 Oct 26. Proc Natl Acad Sci U S A. 2020. PMID: 33106412 Free PMC article.
-
Palaeogenomic analysis of black rat (Rattus rattus) reveals multiple European introductions associated with human economic history.Nat Commun. 2022 May 3;13(1):2399. doi: 10.1038/s41467-022-30009-z. Nat Commun. 2022. PMID: 35504912 Free PMC article.
-
The Justinianic Plague: An inconsequential pandemic?Proc Natl Acad Sci U S A. 2019 Dec 17;116(51):25546-25554. doi: 10.1073/pnas.1903797116. Epub 2019 Dec 2. Proc Natl Acad Sci U S A. 2019. PMID: 31792176 Free PMC article.
-
Environmental Microbial Forensics and Archaeology of Past Pandemics.Microbiol Spectr. 2017 Jan;5(1):10.1128/microbiolspec.emf-0011-2016. doi: 10.1128/microbiolspec.EMF-0011-2016. Microbiol Spectr. 2017. PMID: 28233511 Free PMC article. Review.
References
-
- Benedictow OJ. 2004. The Black Death, 1346-1353: the complete history. England: Boydell & Brewer.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

