A Multi-Serotype Approach Clarifies the Catabolite Control Protein A Regulon in the Major Human Pathogen Group A Streptococcus
- PMID: 27580596
- PMCID: PMC5007534
- DOI: 10.1038/srep32442
A Multi-Serotype Approach Clarifies the Catabolite Control Protein A Regulon in the Major Human Pathogen Group A Streptococcus
Abstract
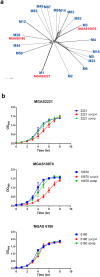
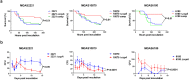
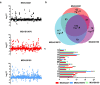
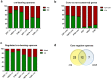
Catabolite control protein A (CcpA) is a highly conserved, master regulator of carbon source utilization in gram-positive bacteria, but the CcpA regulon remains ill-defined. In this study we aimed to clarify the CcpA regulon by determining the impact of CcpA-inactivation on the virulence and transcriptome of three distinct serotypes of the major human pathogen Group A Streptococcus (GAS). CcpA-inactivation significantly decreased GAS virulence in a broad array of animal challenge models consistent with the idea that CcpA is critical to gram-positive bacterial pathogenesis. Via comparative transcriptomics, we established that the GAS CcpA core regulon is enriched for highly conserved CcpA binding motifs (i.e. cre sites). Conversely, strain-specific differences in the CcpA transcriptome seems to consist primarily of affected secondary networks. Refinement of cre site composition via analysis of the core regulon facilitated development of a modified cre consensus that shows promise for improved prediction of CcpA targets in other medically relevant gram-positive pathogens.
Figures

References
-
- Eisenreich W., Dandekar T., Heesemann J. & Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol 8, 401–412 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

