Nonhistone Lysine Methylation in the Regulation of Cancer Pathways
- PMID: 27580749
- PMCID: PMC5088510
- DOI: 10.1101/cshperspect.a026435
Nonhistone Lysine Methylation in the Regulation of Cancer Pathways
Abstract
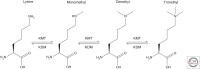
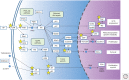
Proteins are regulated by an incredible array of posttranslational modifications (PTMs). Methylation of lysine residues on histone proteins is a PTM with well-established roles in regulating chromatin and epigenetic processes. The recent discovery that hundreds and likely thousands of nonhistone proteins are also methylated at lysine has opened a tremendous new area of research. Major cellular pathways involved in cancer, such as growth signaling and the DNA damage response, are regulated by lysine methylation. Although the field has developed quickly in recent years many fundamental questions remain to be addressed. We review the history and molecular functions of lysine methylation. We then discuss the enzymes that catalyze methylation of lysine residues, the enzymes that remove lysine methylation, and the cancer pathways known to be regulated by lysine methylation. The rest of the article focuses on two open questions that we suggest as a roadmap for future research. First is understanding the large number of candidate methyltransferase and demethylation enzymes whose enzymatic activity is not yet defined and which are potentially associated with cancer through genetic studies. Second is investigating the biological processes and cancer mechanisms potentially regulated by the multitude of lysine methylation sites that have been recently discovered.
Copyright © 2016 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

References
-
- Abu-Farha M, Lanouette S, Elisma F, Tremblay V, Butson J, Figeys D, Couture JF. 2011. Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J Mol Cell Biol 3: 301–308. - PubMed
-
- Ambler RP, Rees MW. 1959. ɛ-N-Methyl-lysine in bacterial flagellar protein. Nature 184: 56–57. - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. - PubMed
-
- Biggar KK, Li SS. 2015. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol 16: 5–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
