High-throughput recombinant protein expression in Escherichia coli: current status and future perspectives
- PMID: 27581654
- PMCID: PMC5008019
- DOI: 10.1098/rsob.160196
High-throughput recombinant protein expression in Escherichia coli: current status and future perspectives
Abstract
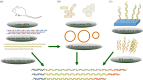
The ease of genetic manipulation, low cost, rapid growth and number of previous studies have made Escherichia coli one of the most widely used microorganism species for producing recombinant proteins. In this post-genomic era, challenges remain to rapidly express and purify large numbers of proteins for academic and commercial purposes in a high-throughput manner. In this review, we describe several state-of-the-art approaches that are suitable for the cloning, expression and purification, conducted in parallel, of numerous molecules, and we discuss recent progress related to soluble protein expression, mRNA folding, fusion tags, post-translational modification and production of membrane proteins. Moreover, we address the ongoing efforts to overcome various challenges faced in protein expression in E. coli, which could lead to an improvement of the current system from trial and error to a predictable and rational design.
Keywords: 5′UTR and N-terminal codons; Escherichia coli; fusion tag; high-throughput; membrane protein; recombinant protein expression.
© 2016 The Authors.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

