ANGIOGENES: knowledge database for protein-coding and noncoding RNA genes in endothelial cells
- PMID: 27582018
- PMCID: PMC5007478
- DOI: 10.1038/srep32475
ANGIOGENES: knowledge database for protein-coding and noncoding RNA genes in endothelial cells
Abstract
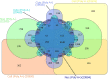
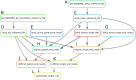
Increasing evidence indicates the presence of long noncoding RNAs (lncRNAs) is specific to various cell types. Although lncRNAs are speculated to be more numerous than protein-coding genes, the annotations of lncRNAs remain primitive due to the lack of well-structured schemes for their identification and description. Here, we introduce a new knowledge database "ANGIOGENES" (http://angiogenes.uni-frankfurt.de) to allow for in silico screening of protein-coding genes and lncRNAs expressed in various types of endothelial cells, which are present in all tissues. Using the latest annotations of protein-coding genes and lncRNAs, publicly-available RNA-seq data was analyzed to identify transcripts that are expressed in endothelial cells of human, mouse and zebrafish. The analyzed data were incorporated into ANGIOGENES to provide a one-stop-shop for transcriptomics data to facilitate further biological validation. ANGIOGENES is an intuitive and easy-to-use database to allow in silico screening of expressed, enriched and/or specific endothelial transcripts under various conditions. We anticipate that ANGIOGENES serves as a starting point for functional studies to elucidate the roles of protein-coding genes and lncRNAs in angiogenesis.
Figures

Similar articles
-
The Proangiogenic Roles of Long NonCoding RNAs Revealed by RNA-Sequencing Following Oxygen-Glucose Deprivation/Re-Oxygenation.Cell Physiol Biochem. 2019;52(4):708-727. doi: 10.33594/000000050. Cell Physiol Biochem. 2019. PMID: 30921509
-
Angiogenic patterning by STEEL, an endothelial-enriched long noncoding RNA.Proc Natl Acad Sci U S A. 2018 Mar 6;115(10):2401-2406. doi: 10.1073/pnas.1715182115. Epub 2018 Feb 21. Proc Natl Acad Sci U S A. 2018. PMID: 29467285 Free PMC article.
-
zflncRNApedia: A Comprehensive Online Resource for Zebrafish Long Non-Coding RNAs.PLoS One. 2015 Jun 11;10(6):e0129997. doi: 10.1371/journal.pone.0129997. eCollection 2015. PLoS One. 2015. PMID: 26065909 Free PMC article.
-
The Working Modules of Long Noncoding RNAs in Cancer Cells.Adv Exp Med Biol. 2016;927:49-67. doi: 10.1007/978-981-10-1498-7_2. Adv Exp Med Biol. 2016. PMID: 27376731 Review.
-
The specificity of long noncoding RNA expression.Biochim Biophys Acta. 2016 Jan;1859(1):16-22. doi: 10.1016/j.bbagrm.2015.08.005. Epub 2015 Aug 19. Biochim Biophys Acta. 2016. PMID: 26297315 Review.
Cited by
-
Magnetique: an interactive web application to explore transcriptome signatures of heart failure.J Transl Med. 2022 Nov 7;20(1):513. doi: 10.1186/s12967-022-03694-z. J Transl Med. 2022. PMID: 36345035 Free PMC article.
-
Computational analysis of ribonomics datasets identifies long non-coding RNA targets of γ-herpesviral miRNAs.Nucleic Acids Res. 2018 Sep 19;46(16):8574-8589. doi: 10.1093/nar/gky459. Nucleic Acids Res. 2018. PMID: 29846699 Free PMC article.
-
Using Human iPSC-Derived Neurons to Uncover Activity-Dependent Non-Coding RNAs.Genes (Basel). 2017 Dec 20;8(12):401. doi: 10.3390/genes8120401. Genes (Basel). 2017. PMID: 29261115 Free PMC article.
-
The Regulatory Roles of Non-coding RNAs in Angiogenesis and Neovascularization From an Epigenetic Perspective.Front Oncol. 2019 Oct 24;9:1091. doi: 10.3389/fonc.2019.01091. eCollection 2019. Front Oncol. 2019. PMID: 31709179 Free PMC article. Review.
-
Pan-tissue transcriptome analysis of long noncoding RNAs in the American beaver Castor canadensis.BMC Genomics. 2020 Feb 12;21(1):153. doi: 10.1186/s12864-019-6432-4. BMC Genomics. 2020. PMID: 32050897 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

