Proteome-wide association studies identify biochemical modules associated with a wing-size phenotype in Drosophila melanogaster
- PMID: 27582081
- PMCID: PMC5025782
- DOI: 10.1038/ncomms12649
Proteome-wide association studies identify biochemical modules associated with a wing-size phenotype in Drosophila melanogaster
Abstract
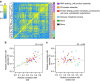
The manner by which genetic diversity within a population generates individual phenotypes is a fundamental question of biology. To advance the understanding of the genotype-phenotype relationships towards the level of biochemical processes, we perform a proteome-wide association study (PWAS) of a complex quantitative phenotype. We quantify the variation of wing imaginal disc proteomes in Drosophila genetic reference panel (DGRP) lines using SWATH mass spectrometry. In spite of the very large genetic variation (1/36 bp) between the lines, proteome variability is surprisingly small, indicating strong molecular resilience of protein expression patterns. Proteins associated with adult wing size form tight co-variation clusters that are enriched in fundamental biochemical processes. Wing size correlates with some basic metabolic functions, positively with glucose metabolism but negatively with mitochondrial respiration and not with ribosome biogenesis. Our study highlights the power of PWAS to filter functional variants from the large genetic variability in natural populations.
Figures





References
-
- Edgar B. A. How flies get their size: genetics meets physiology. Nat. Rev. Genet. 7, 907–916 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

