In silico identification of enhancers on the basis of a combination of transcription factor binding motif occurrences
- PMID: 27582178
- PMCID: PMC5007594
- DOI: 10.1038/srep32476
In silico identification of enhancers on the basis of a combination of transcription factor binding motif occurrences
Abstract
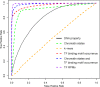
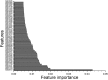
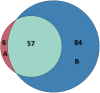
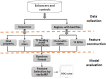
Enhancers interact with gene promoters and form chromatin looping structures that serve important functions in various biological processes, such as the regulation of gene transcription and cell differentiation. However, enhancers are difficult to identify because they generally do not have fixed positions or consensus sequence features, and biological experiments for enhancer identification are costly in terms of labor and expense. In this work, several models were built by using various sequence-based feature sets and their combinations for enhancer prediction. The selected features derived from a recursive feature elimination method showed that the model using a combination of 141 transcription factor binding motif occurrences from 1,422 transcription factor position weight matrices achieved a favorably high prediction accuracy superior to that of other reported methods. The models demonstrated good prediction accuracy for different enhancer datasets obtained from different cell lines/tissues. In addition, prediction accuracy was further improved by integration of chromatin state features. Our method is complementary to wet-lab experimental methods and provides an additional method to identify enhancers.
Figures


Similar articles
-
Enhancer prediction in the human genome by probabilistic modelling of the chromatin feature patterns.BMC Bioinformatics. 2020 Jul 20;21(1):317. doi: 10.1186/s12859-020-03621-3. BMC Bioinformatics. 2020. PMID: 32689977 Free PMC article.
-
Genome-wide prediction of transcription factor binding sites using an integrated model.Genome Biol. 2010 Jan 22;11(1):R7. doi: 10.1186/gb-2010-11-1-r7. Genome Biol. 2010. PMID: 20096096 Free PMC article.
-
Role of chromatin and transcriptional co-regulators in mediating p63-genome interactions in keratinocytes.BMC Genomics. 2014 Nov 29;15(1):1042. doi: 10.1186/1471-2164-15-1042. BMC Genomics. 2014. PMID: 25433490 Free PMC article.
-
Evaluating Enhancer Function and Transcription.Annu Rev Biochem. 2020 Jun 20;89:213-234. doi: 10.1146/annurev-biochem-011420-095916. Epub 2020 Mar 20. Annu Rev Biochem. 2020. PMID: 32197056 Review.
-
Eukaryotic enhancers: common features, regulation, and participation in diseases.Cell Mol Life Sci. 2015 Jun;72(12):2361-75. doi: 10.1007/s00018-015-1871-9. Epub 2015 Feb 26. Cell Mol Life Sci. 2015. PMID: 25715743 Free PMC article. Review.
Cited by
-
iEnhancer-ECNN: identifying enhancers and their strength using ensembles of convolutional neural networks.BMC Genomics. 2019 Dec 24;20(Suppl 9):951. doi: 10.1186/s12864-019-6336-3. BMC Genomics. 2019. PMID: 31874637 Free PMC article.
-
Integrative analysis of transcriptomic and epigenomic data reveals distinct patterns for developmental and housekeeping gene regulation.BMC Biol. 2024 Apr 10;22(1):78. doi: 10.1186/s12915-024-01869-2. BMC Biol. 2024. PMID: 38600550 Free PMC article.
-
DNA Methylation of Enhancer Elements in Myeloid Neoplasms: Think Outside the Promoters?Cancers (Basel). 2019 Sep 24;11(10):1424. doi: 10.3390/cancers11101424. Cancers (Basel). 2019. PMID: 31554341 Free PMC article. Review.
-
Three-dimensional texture features from intensity and high-order derivative maps for the discrimination between bladder tumors and wall tissues via MRI.Int J Comput Assist Radiol Surg. 2017 Apr;12(4):645-656. doi: 10.1007/s11548-017-1522-8. Epub 2017 Jan 21. Int J Comput Assist Radiol Surg. 2017. PMID: 28110476
-
Epigenomic landscape of enhancer elements during Hydra head organizer formation.Epigenetics Chromatin. 2020 Oct 12;13(1):43. doi: 10.1186/s13072-020-00364-6. Epigenetics Chromatin. 2020. PMID: 33046126 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

