Adaptive molecular evolution of the two-pore channel 1 gene TPC1 in the karst-adapted genus Primulina (Gesneriaceae)
- PMID: 27582362
- PMCID: PMC5155596
- DOI: 10.1093/aob/mcw168
Adaptive molecular evolution of the two-pore channel 1 gene TPC1 in the karst-adapted genus Primulina (Gesneriaceae)
Abstract
Background and aims: Limestone karst areas possess high floral diversity and endemism. The genus Primulina, which contributes to the unique calcicole flora, has high species richness and exhibit specific soil-based habitat associations that are mainly distributed on calcareous karst soils. The adaptive molecular evolutionary mechanism of the genus to karst calcium-rich environments is still not well understood. The Ca2+-permeable channel TPC1 was used in this study to test whether its gene is involved in the local adaptation of Primulina to karst high-calcium soil environments.
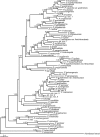
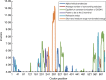
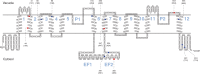
Methods: Specific amplification and sequencing primers were designed and used to amplify the full-length coding sequences of TPC1 from cDNA of 76 Primulina species. The sequence alignment without recombination and the corresponding reconstructed phylogeny tree were used in molecular evolutionary analyses at the nucleic acid level and amino acid level, respectively. Finally, the identified sites under positive selection were labelled on the predicted secondary structure of TPC1.
Key results: Seventy-six full-length coding sequences of Primulina TPC1 were obtained. The length of the sequences varied between 2220 and 2286 bp and the insertion/deletion was located at the 5' end of the sequences. No signal of substitution saturation was detected in the sequences, while significant recombination breakpoints were detected. The molecular evolutionary analyses showed that TPC1 was dominated by purifying selection and the selective pressures were not significantly different among species lineages. However, significant signals of positive selection were detected at both TPC1 codon level and amino acid level, and five sites under positive selective pressure were identified by at least three different methods.
Conclusions: The Ca2+-permeable channel TPC1 may be involved in the local adaptation of Primulina to karst Ca2+-rich environments. Different species lineages suffered similar selective pressure associated with calcium in karst environments, and episodic diversifying selection at a few sites may play a major role in the molecular evolution of Primulina TPC1.
Keywords: Ca2+-permeable channel; Primulina; TPC1; calcium; karst; molecular evolution; positive selection.
© The Author 2016. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
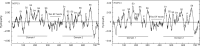
Similar articles
-
Adaptive Molecular Evolution of PHYE in Primulina, a Karst Cave Plant.PLoS One. 2015 Jun 1;10(6):e0127821. doi: 10.1371/journal.pone.0127821. eCollection 2015. PLoS One. 2015. PMID: 26030408 Free PMC article.
-
Plastomes of limestone karst gesneriad genera Petrocodon and Primulina, and the comparative plastid phylogenomics of Gesneriaceae.Sci Rep. 2022 Sep 22;12(1):15800. doi: 10.1038/s41598-022-19812-2. Sci Rep. 2022. PMID: 36138079 Free PMC article.
-
Nitrogen limitation as a driver of genome size evolution in a group of karst plants.Sci Rep. 2015 Jun 25;5:11636. doi: 10.1038/srep11636. Sci Rep. 2015. PMID: 26109237 Free PMC article.
-
Five new species of the genus Primulina (Gesneriaceae) from Limestone Areas of Guangxi Zhuangzu Autonomous Region, China.PhytoKeys. 2019 Jul 19;127:77-91. doi: 10.3897/phytokeys.127.35445. eCollection 2019. PhytoKeys. 2019. PMID: 31379451 Free PMC article. Review.
-
Structure and Function of TPC1 Vacuole SV Channel Gains Shape.Mol Plant. 2018 Jun 4;11(6):764-775. doi: 10.1016/j.molp.2018.03.017. Epub 2018 Mar 31. Mol Plant. 2018. PMID: 29614320 Review.
Cited by
-
Comprehensive Comparative Analyses of Aspidistra Chloroplast Genomes: Insights into Interspecific Plastid Diversity and Phylogeny.Genes (Basel). 2023 Sep 29;14(10):1894. doi: 10.3390/genes14101894. Genes (Basel). 2023. PMID: 37895243 Free PMC article.
-
Plant adaptability in karst regions.J Plant Res. 2021 Sep;134(5):889-906. doi: 10.1007/s10265-021-01330-3. Epub 2021 Jul 13. J Plant Res. 2021. PMID: 34258691 Review.
-
A Combined Morphological and Molecular Evolutionary Analysis of Karst-Environment Adaptation for the Genus Urophysa (Ranunculaceae).Front Plant Sci. 2021 Jun 10;12:667988. doi: 10.3389/fpls.2021.667988. eCollection 2021. Front Plant Sci. 2021. PMID: 34177982 Free PMC article.
-
Genomic Insights into Adaptation to Karst Limestone and Incipient Speciation in East Asian Platycarya spp. (Juglandaceae).Mol Biol Evol. 2023 Jun 1;40(6):msad121. doi: 10.1093/molbev/msad121. Mol Biol Evol. 2023. PMID: 37216901 Free PMC article.
-
Comparative Analysis of the Chloroplast Genomes of Cypripedium: Assessing the Roles of SSRs and TRs in the Non-Coding Regions of LSC in Shaping Chloroplast Genome Size.Int J Mol Sci. 2025 Apr 14;26(8):3691. doi: 10.3390/ijms26083691. Int J Mol Sci. 2025. PMID: 40332341 Free PMC article.
References
-
- Ai B, Gao Y, Zhang XL, Tao JJ, Kang M, Huang HW. 2015. Comparative transcriptome resources of eleven Primulina species, a group of ‘stone plants' from a biodiversity hot spot. Molecular Ecology Resources 15: 619–632. - PubMed
-
- Barkman TJ, Martins TR, Sutton E, Stout JT. 2007. Positive selection for single amino acid change promotes substrate discrimination of a plant volatile-producing enzyme. Molecular Biology and Evolution 24: 1320–1329. - PubMed
-
- Bielawski JP, Yang ZH. 2003. Maximum likelihood methods for detecting adaptive evolution after gene duplication. Journal of Structural and Functional Genomics 3: 201–212. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

