Clus-DoC: a combined cluster detection and colocalization analysis for single-molecule localization microscopy data
- PMID: 27582387
- PMCID: PMC5221594
- DOI: 10.1091/mbc.E16-07-0478
Clus-DoC: a combined cluster detection and colocalization analysis for single-molecule localization microscopy data
Abstract
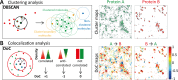
Advances in fluorescence microscopy are providing increasing evidence that the spatial organization of proteins in cell membranes may facilitate signal initiation and integration for appropriate cellular responses. Our understanding of how changes in spatial organization are linked to function has been hampered by the inability to directly measure signaling activity or protein association at the level of individual proteins in intact cells. Here we solve this measurement challenge by developing Clus-DoC, an analysis strategy that quantifies both the spatial distribution of a protein and its colocalization status. We apply this approach to the triggering of the T-cell receptor during T-cell activation, as well as to the functionality of focal adhesions in fibroblasts, thereby demonstrating an experimental and analytical workflow that can be used to quantify signaling activity and protein colocalization at the level of individual proteins.
© 2016 Pageon et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures





References
-
- Ankerst M, Breunig MM, Kriegel H-P, Sander J. OPTICS: ordering points to identify the clustering structure. ACM SIGMOD Record. 1999;28:49–60.
-
- Annibale P, Scarselli M, Kodiyan A, Radenovic A. Photoactivatable fluorescent protein mEos2 displays repeated photoactivation after a long-lived dark state in the red photoconverted form. J Phys Chem Lett. 2010;1:1506–1510.
-
- Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Identification of clustering artifacts in photoactivated localization microscopy. Nat Methods. 2011;8:527–528. - PubMed
-
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

