A microchip platform for structural oncology applications
- PMID: 27583302
- PMCID: PMC5003533
- DOI: 10.1038/npjbcancer.2016.16
A microchip platform for structural oncology applications
Abstract
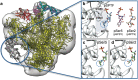
Recent advances in the development of functional materials offer new tools to dissect human health and disease mechanisms. The use of tunable surfaces is especially appealing as substrates can be tailored to fit applications involving specific cell types or tissues. Here we use tunable materials to facilitate the three-dimensional (3D) analysis of BRCA1 gene regulatory complexes derived from human cancer cells. We employed a recently developed microchip platform to isolate BRCA1 protein assemblies natively formed in breast cancer cells with and without BRCA1 mutations. The captured assemblies proved amenable to cryo-electron microscopy (EM) imaging and downstream computational analysis. Resulting 3D structures reveal the manner in which wild-type BRCA1 engages the RNA polymerase II (RNAP II) core complex that contained K63-linked ubiquitin moieties-a putative signal for DNA repair. Importantly, we also determined that molecular assemblies harboring the BRCA15382insC mutation exhibited altered protein interactions and ubiquitination patterns compared to wild-type complexes. Overall, our analyses proved optimal for developing new structural oncology applications involving patient-derived cancer cells, while expanding our knowledge of BRCA1's role in gene regulatory events.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Le Page, F. et al. BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Res. 60, 5548–5552 (2000). - PubMed
-
- King, M. C., Marks, J. H. & Mandell, J. B. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302, 643–646 (2003). - PubMed
-
- Caestecker, K. W. & Van de Walle, G. R. The role of BRCA1 in DNA double-strand repair: past and present. Exp. Cell Res. 319, 575–587 (2013). - PubMed
-
- Friedman, L. S. et al. The search for BRCA1. Cancer Res. 54, 6374–6382 (1994). - PubMed
-
- Wu, L. C. et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 14, 430–440 (1996). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

