HSV-1-induced activation of NF-κB protects U937 monocytic cells against both virus replication and apoptosis
- PMID: 27584793
- PMCID: PMC5059854
- DOI: 10.1038/cddis.2016.250
HSV-1-induced activation of NF-κB protects U937 monocytic cells against both virus replication and apoptosis
Abstract
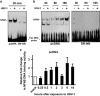
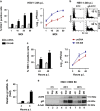
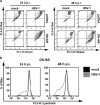
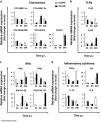
The transcription factor nuclear factor-kappa B (NF-κB) is a crucial player of the antiviral innate response. Intriguingly, however, NF-κB activation is assumed to favour herpes simplex virus (HSV) infection rather than restrict it. Apoptosis, a form of innate response to viruses, is completely inhibited by HSV in fully permissive cells, but not in cells incapable to fully sustain HSV replication, such as immunocompetent cells. To resolve the intricate interplay among NF-κB signalling, apoptosis and permissiveness to HSV-1 in monocytic cells, we utilized U937 monocytic cells in which NF-κB activation was inhibited by expressing a dominant-negative IκBα. Surprisingly, viral production was increased in monocytic cells in which NF-κB was inhibited. Moreover, inhibition of NF-κB led to increased apoptosis following HSV-1 infection, associated with lysosomal membrane permeabilization. High expression of late viral proteins and induction of apoptosis occurred in distinct cells. Transcriptional analysis of known innate response genes by real-time quantitative reverse transcription-PCR excluded a contribution of the assayed genes to the observed phenomena. Thus, in monocytic cells NF-κB activation simultaneously serves as an innate process to restrict viral replication as well as a mechanism to limit the damage of an excessive apoptotic response to HSV-1 infection. This finding may clarify mechanisms controlling HSV-1 infection in monocytic cells.
Figures





References
-
- Hiscott J. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev 2007; 18: 483–490. - PubMed
-
- Le Negrate G. Viral interference with innate immunity by preventing NF-kappaB activity. Cell Microbiol 2012; 14: 168–181. - PubMed
-
- Roizman B, Knipe DM, Whitley RJ. Herpes simplex virus. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA et al (eds). Fields Virology, 6th edn. vol. 2. Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013, pp 1823–1897.
-
- Rong BL, Libermann TA, Kogawa K, Ghosh S, Cao LX, Pavanlangston D et al. Hsv-1-inducible proteins bind to Nf-kappa-B-like sites in the Hsv-1 genome. Virology 1992; 189: 750–756. - PubMed
-
- Medici MA, Sciortino MT, Perri D, Amici C, Avitabile E, Ciotti M et al. Protection by herpes simplex virus glycoprotein D against Fas-mediated apoptosis: role of nuclear factor kappaB. J Biol Chem 2003; 278: 36059–36067. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

