Rare genetic variation in UNC13A may modify survival in amyotrophic lateral sclerosis
- PMID: 27584932
- PMCID: PMC5125285
- DOI: 10.1080/21678421.2016.1213852
Rare genetic variation in UNC13A may modify survival in amyotrophic lateral sclerosis
Abstract
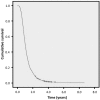
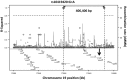
Our objective was to identify whether rare genetic variation in amyotrophic lateral sclerosis (ALS) candidate survival genes modifies ALS survival. Candidate genes were selected based on evidence for modifying ALS survival. Each tail of the extreme 1.5% of survival was selected from the UK MND DNA Bank and all samples available underwent whole genome sequencing. A replication set from the Netherlands was used for validation. Sequences of candidate survival genes were extracted and variants passing quality control with a minor allele frequency ≤0.05 were selected for association testing. Analysis was by burden testing using SKAT. Candidate survival genes UNC13A, KIFAP3, and EPHA4 were tested for association in a UK sample comprising 25 short survivors and 25 long survivors. Results showed that only SNVs in UNC13A were associated with survival (p = 6.57 × 10-3). SNV rs10419420:G > A was found exclusively in long survivors (3/25) and rs4808092:G > A exclusively in short survivors (4/25). These findings were not replicated in a Dutch sample. In conclusion, population specific rare variants of UNC13A may modulate survival in ALS.
Keywords: ALS; UNC13A; genetic modifiers; sequencing; survival.
Figures
Similar articles
-
Genetic factors for survival in amyotrophic lateral sclerosis: an integrated approach combining a systematic review, pairwise and network meta-analysis.BMC Med. 2022 Jun 27;20(1):209. doi: 10.1186/s12916-022-02411-3. BMC Med. 2022. PMID: 35754054 Free PMC article.
-
Association of UNC13A with increased amyotrophic lateral sclerosis risk, bulbar onset, and lower motor neuron involvement in a Norwegian ALS cohort.Amyotroph Lateral Scler Frontotemporal Degener. 2025 Aug;26(5-6):566-572. doi: 10.1080/21678421.2024.2447922. Epub 2024 Dec 30. Amyotroph Lateral Scler Frontotemporal Degener. 2025. PMID: 40135631
-
Mechanical ventilation for amyotrophic lateral sclerosis/motor neuron disease.Cochrane Database Syst Rev. 2017 Oct 6;10(10):CD004427. doi: 10.1002/14651858.CD004427.pub4. Cochrane Database Syst Rev. 2017. PMID: 28982219 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Treatment for sialorrhea (excessive saliva) in people with motor neuron disease/amyotrophic lateral sclerosis.Cochrane Database Syst Rev. 2022 May 20;5(5):CD006981. doi: 10.1002/14651858.CD006981.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593746 Free PMC article.
Cited by
-
The era of cryptic exons: implications for ALS-FTD.Mol Neurodegener. 2023 Mar 15;18(1):16. doi: 10.1186/s13024-023-00608-5. Mol Neurodegener. 2023. PMID: 36922834 Free PMC article. Review.
-
Meta-analysis of pharmacogenetic interactions in amyotrophic lateral sclerosis clinical trials.Neurology. 2017 Oct 31;89(18):1915-1922. doi: 10.1212/WNL.0000000000004606. Epub 2017 Oct 4. Neurology. 2017. PMID: 28978660 Free PMC article. Review.
-
Biomarkers in Motor Neuron Disease: A State of the Art Review.Front Neurol. 2019 Apr 3;10:291. doi: 10.3389/fneur.2019.00291. eCollection 2019. Front Neurol. 2019. PMID: 31001186 Free PMC article. Review.
-
What causes amyotrophic lateral sclerosis?F1000Res. 2017 Mar 28;6:371. doi: 10.12688/f1000research.10476.1. eCollection 2017. F1000Res. 2017. PMID: 28408982 Free PMC article. Review.
-
Linkage of Alzheimer disease families with Puerto Rican ancestry identifies a chromosome 9 locus.Neurobiol Aging. 2021 Aug;104:115.e1-115.e7. doi: 10.1016/j.neurobiolaging.2021.02.019. Epub 2021 Feb 28. Neurobiol Aging. 2021. PMID: 33902942 Free PMC article.
References
-
- Al-Chalabi A, Hardiman O. The epidemiology of ALS: a conspiracy of genes, environment and time. Nature reviews. Neurology. 2013;9:617–28. - PubMed
-
- Hardiman O, van den Berg LH, Kiernan MC. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nature reviews. Neurology. 2011;7:639–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
