Functions and Malfunctions of Mammalian DNA-Cytosine Deaminases
- PMID: 27585283
- PMCID: PMC5528147
- DOI: 10.1021/acs.chemrev.6b00296
Functions and Malfunctions of Mammalian DNA-Cytosine Deaminases
Abstract
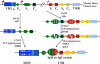
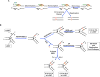
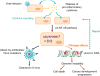
The AID/APOBEC family enzymes convert cytosines in single-stranded DNA to uracils, causing base substitutions and strand breaks. They are induced by cytokines produced during the body's inflammatory response to infections, and they help combat infections through diverse mechanisms. AID is essential for the maturation of antibodies and causes mutations and deletions in antibody genes through somatic hypermutation (SHM) and class-switch recombination (CSR) processes. One member of the APOBEC family, APOBEC1, edits mRNA for a protein involved in lipid transport. Members of the APOBEC3 subfamily in humans (APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H) inhibit infections of viruses such as HIV-1, HBV, and HCV, and retrotransposition of endogenous retroelements through mutagenic and nonmutagenic mechanisms. There is emerging consensus that these enzymes can cause mutations in the cellular genome at replication forks or within transcription bubbles depending on the physiological state of the cell and the phase of the cell cycle during which they are expressed. We describe here the state of knowledge about the structures of these enzymes, regulation of their expression, and both the advantageous and deleterious consequences of their expression, including carcinogenesis. We highlight similarities among them and present a holistic view of their regulation and function.
Figures

References
-
- Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. Journal of molecular biology. 2004;337:585. - PubMed
-
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726. - PubMed
-
- Petersen-Mahrt SK, Neuberger MS. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1) The Journal of biological chemistry. 2003;278:19583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

