Towards understanding brain-gut-microbiome connections in Alzheimer's disease
- PMID: 27585440
- PMCID: PMC5009560
- DOI: 10.1186/s12918-016-0307-y
Towards understanding brain-gut-microbiome connections in Alzheimer's disease
Abstract
Background: Alzheimer's disease (AD) is complex, with genetic, epigenetic, and environmental factors contributing to disease susceptibility and progression. While significant progress has been made in understanding genetic, molecular, behavioral, and neurological aspects of AD, relatively little is known about which environmental factors are important in AD etiology and how they interact with genetic factors in the development of AD. Here, we propose a data-driven, hypotheses-free computational approach to characterize which and how human gut microbial metabolites, an important modifiable environmental factor, may contribute to various aspects of AD.
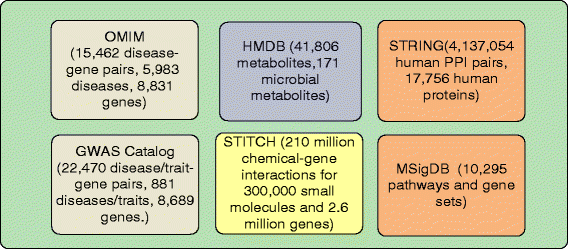
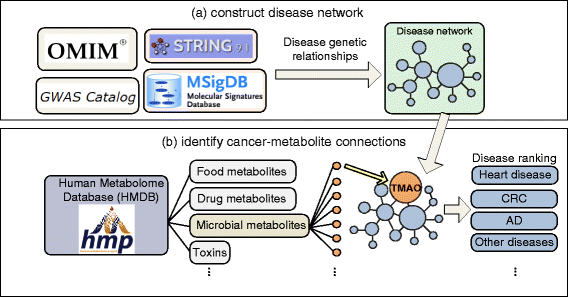
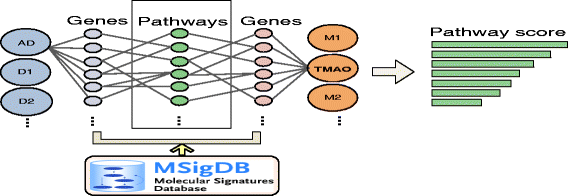
Materials and methods: We integrated vast amounts of complex and heterogeneous biomedical data, including disease genetics, chemical genetics, human microbial metabolites, protein-protein interactions, and genetic pathways. We developed a novel network-based approach to model the genetic interactions between all human microbial metabolites and genetic diseases. We identified metabolites that share significant genetic commonality with AD in humans. We developed signal prioritization algorithms to identify the co-regulated genetic pathways underlying the identified AD-metabolite (brain-gut) connections.
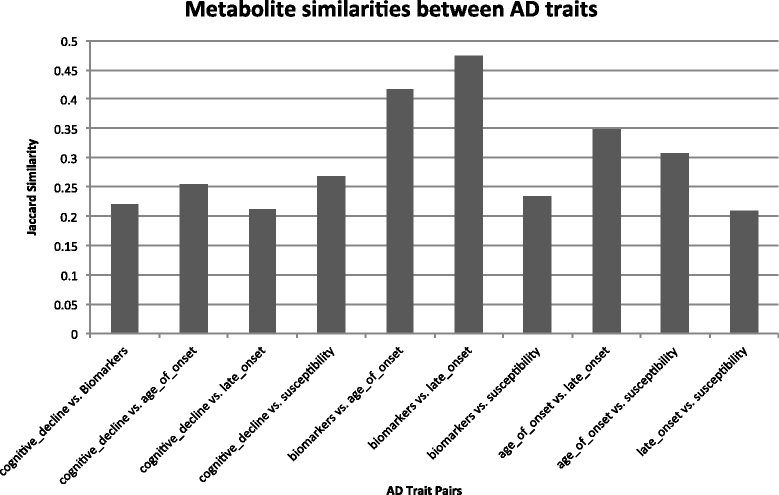
Results: We validated our algorithms using known microbial metabolite-AD associations, namely AD-3,4-dihydroxybenzeneacetic acid, AD-mannitol, and AD-succinic acid. Our study provides supporting evidence that human gut microbial metabolites may be an important mechanistic link between environmental exposure and various aspects of AD. We identified metabolites that are significantly associated with various aspects in AD, including AD susceptibility, cognitive decline, biomarkers, age of onset, and the onset of AD. We identified common genetic pathways underlying AD biomarkers and its top one ranked metabolite trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. These coregulated pathways between TMAO-AD may provide insights into the mechanisms of how dietary meat and fat contribute to AD.
Conclusions: Employing an integrated computational approach, we provide intriguing and supporting evidence for a role of microbial metabolites, an important modifiable environmental factor, in AD etiology. Our study provides the foundations for subsequent hypothesis-driven biological and clinical studies of brain-gut-environment interactions in AD.
Keywords: Alzheimer’s disease; Disease etiology; Human gut micriobiome; Network medicine; Systems biology.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

