Bioinformatic analysis of genotype by sequencing (GBS) data with NGSEP
- PMID: 27585926
- PMCID: PMC5009557
- DOI: 10.1186/s12864-016-2827-7
Bioinformatic analysis of genotype by sequencing (GBS) data with NGSEP
Abstract
Background: Therecent development and availability of different genotype by sequencing (GBS) protocols provided a cost-effective approach to perform high-resolution genomic analysis of entire populations in different species. The central component of all these protocols is the digestion of the initial DNA with known restriction enzymes, to generate sequencing fragments at predictable and reproducible sites. This allows to genotype thousands of genetic markers on populations with hundreds of individuals. Because GBS protocols achieve parallel genotyping through high throughput sequencing (HTS), every GBS protocol must include a bioinformatics pipeline for analysis of HTS data. Our bioinformatics group recently developed the Next Generation Sequencing Eclipse Plugin (NGSEP) for accurate, efficient, and user-friendly analysis of HTS data.
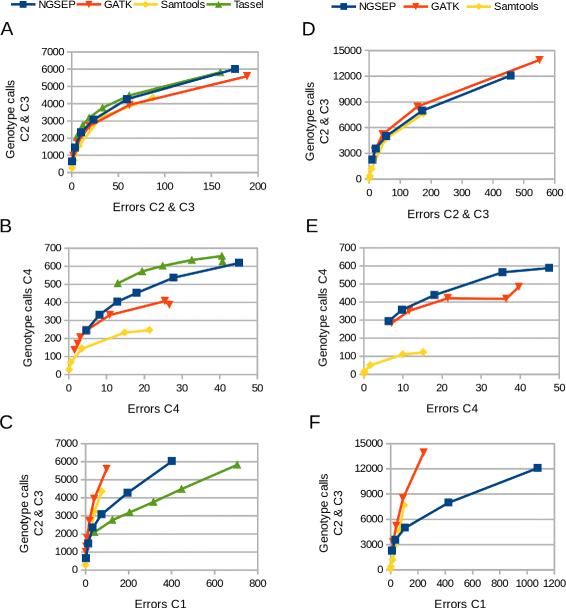
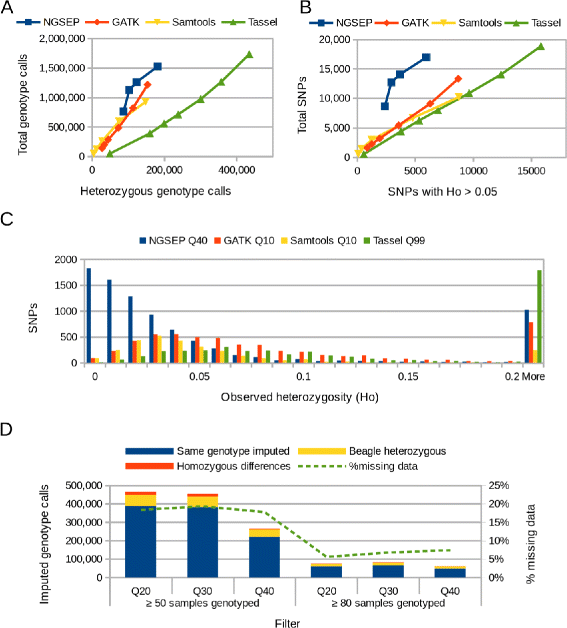
Results: Here we present the latest functionalities implemented in NGSEP in the context of the analysis of GBS data. We implemented a one step wizard to perform parallel read alignment, variants identification and genotyping from HTS reads sequenced from entire populations. We added different filters for variants, samples and genotype calls as well as calculation of summary statistics overall and per sample, and diversity statistics per site. NGSEP includes a module to translate genotype calls to some of the most widely used input formats for integration with several tools to perform downstream analyses such as population structure analysis, construction of genetic maps, genetic mapping of complex traits and phenotype prediction for genomic selection. We assessed the accuracy of NGSEP on two highly heterozygous F1 cassava populations and on an inbred common bean population, and we showed that NGSEP provides similar or better accuracy compared to other widely used software packages for variants detection such as GATK, Samtools and Tassel.
Conclusions: NGSEP is a powerful, accurate and efficient bioinformatics software tool for analysis of HTS data, and also one of the best bioinformatic packages to facilitate the analysis and to maximize the genomic variability information that can be obtained from GBS experiments for population genomics.
Keywords: Bioinformatics; GBS; NGSEP; SNP calling; Sequencing.
Figures


References
-
- Spindel J, Begum H, Akdemir D, Virk P, Collard B, Redoña E, et al. Genomic selection and association mapping in rice (Oryza sativa): effect of trait genetic architecture, training population composition, marker number and statistical model on accuracy of rice genomic selection in elite, tropical rice breeding lines. PLoS Genet. 2015;11(2):e1004982. doi: 10.1371/journal.pgen.1004982. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

