Comparison of blood RNA isolation methods from samples stabilized in Tempus tubes and stored at a large human biobank
- PMID: 27587079
- PMCID: PMC5009671
- DOI: 10.1186/s13104-016-2224-y
Comparison of blood RNA isolation methods from samples stabilized in Tempus tubes and stored at a large human biobank
Abstract
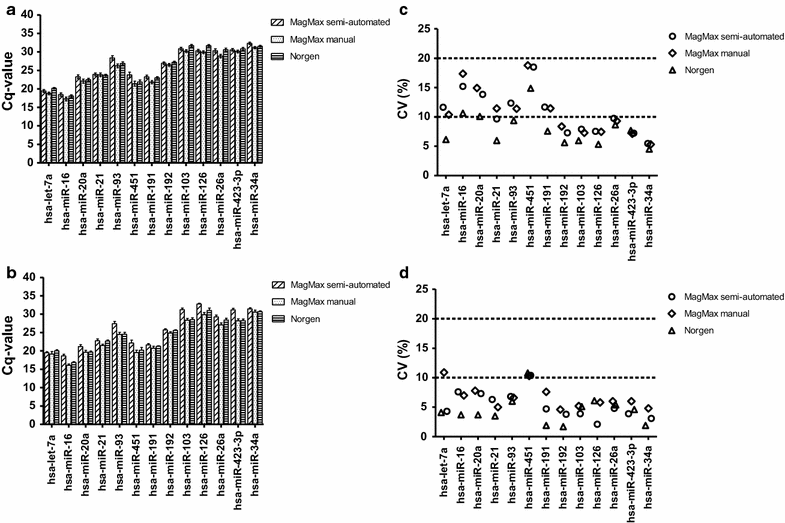
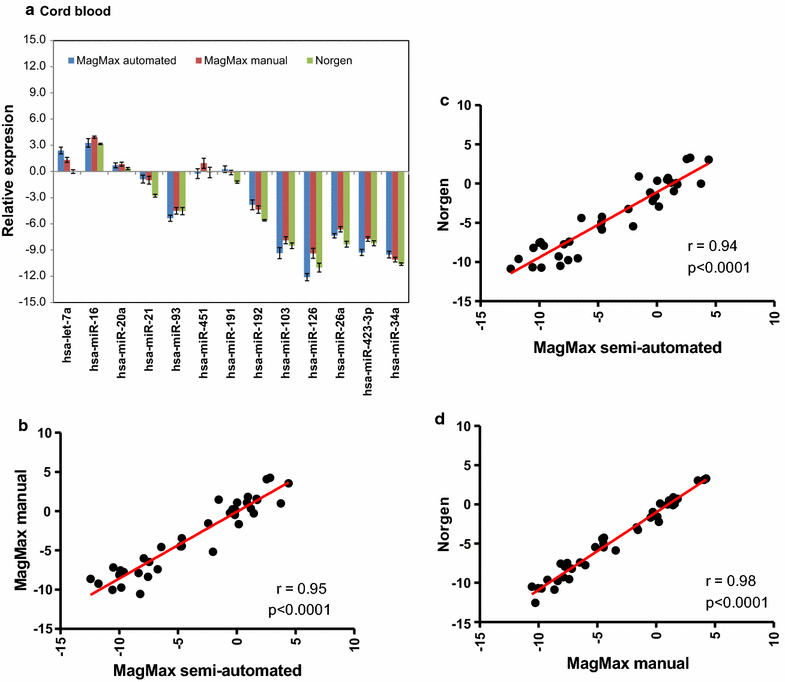
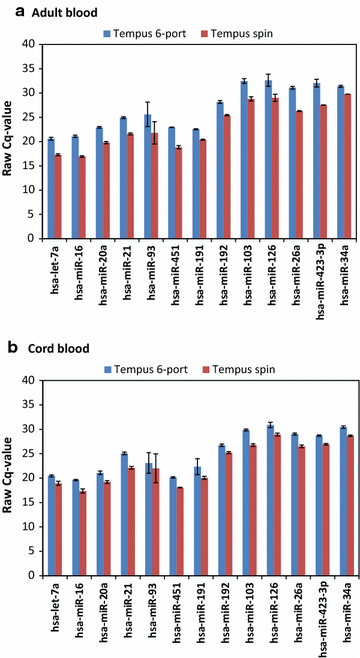
Background: More than 50,000 adult and cord blood samples were collected in Tempus tubes and stored at the Norwegian Institute of Public Health Biobank for future use. In this study, we systematically evaluated and compared five blood-RNA isolation protocols: three blood-RNA isolation protocols optimized for simultaneous isolation of all blood-RNA species (MagMAX RNA Isolation Kit, both manual and semi-automated protocols; and Norgen Preserved Blood RNA kit I); and two protocols optimized for large RNAs only (Tempus Spin RNA, and Tempus 6-port isolation kit). We estimated the following parameters: RNA quality, RNA yield, processing time, cost per sample, and RNA transcript stability of six selected mRNAs and 13 miRNAs using real-time qPCR.
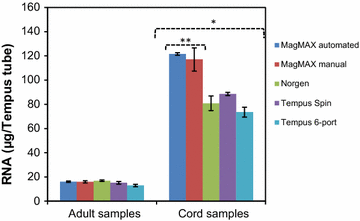
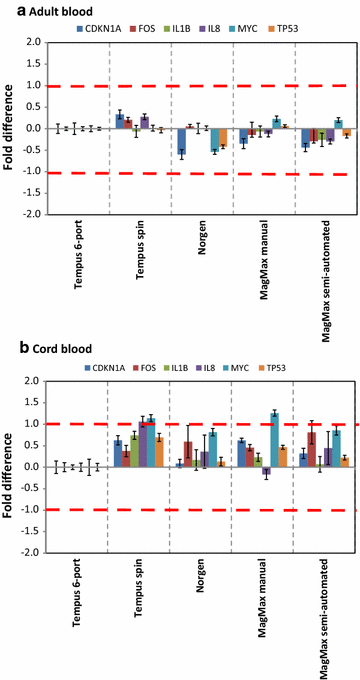
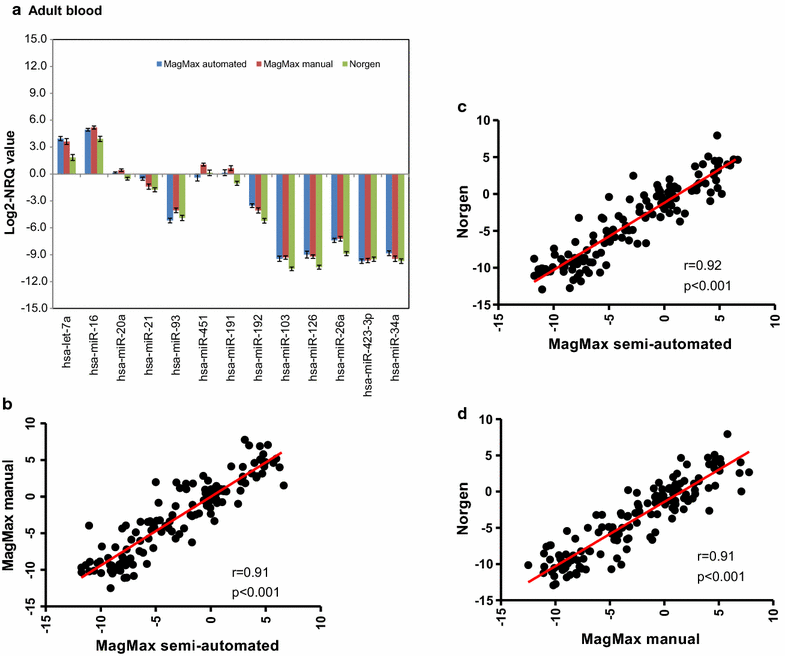
Findings: Whole blood samples from adults (n = 59 tubes) and umbilical cord blood (n = 18 tubes) samples collected in Tempus tubes were analyzed. High-quality blood-RNAs with average RIN-values above seven were extracted using all five RNA isolation protocols. The transcript levels of the six selected genes showed minimal variation between the five protocols. Unexplained differences within the transcript levels of the 13 miRNA were observed; however, the 13 miRNAs had similar expression direction and they were within the same order of magnitude. Some differences in the RNA processing time and cost were noted.
Conclusions: Sufficient amounts of high-quality RNA were obtained using all five protocols, and the Tempus blood RNA system therefore seems not to be dependent on one specific RNA isolation method.
Keywords: Biobank; Cord blood; Epigenetics; Gene expression; MiRNA; MoBa; Noncoding RNA; RNA isolation; Tempus tubes; The Norwegian Mother and Child Cohort Study.
Figures
Similar articles
-
Simultaneous extraction of mRNA and microRNA from whole blood stabilized in tempus tubes.BMC Res Notes. 2019 Jan 18;12(1):39. doi: 10.1186/s13104-019-4087-5. BMC Res Notes. 2019. PMID: 30658701 Free PMC article.
-
Comparison of whole blood RNA preservation tubes and novel generation RNA extraction kits for analysis of mRNA and MiRNA profiles.PLoS One. 2014 Dec 3;9(12):e113298. doi: 10.1371/journal.pone.0113298. eCollection 2014. PLoS One. 2014. PMID: 25469788 Free PMC article.
-
Long-term storage of blood RNA collected in RNA stabilizing Tempus tubes in a large biobank--evaluation of RNA quality and stability.BMC Res Notes. 2014 Sep 12;7:633. doi: 10.1186/1756-0500-7-633. BMC Res Notes. 2014. PMID: 25214016 Free PMC article.
-
MicroRNA isolation and stability in stored RNA samples.Biochem Biophys Res Commun. 2009 Dec 4;390(1):1-4. doi: 10.1016/j.bbrc.2009.09.061. Epub 2009 Sep 19. Biochem Biophys Res Commun. 2009. PMID: 19769940 Review.
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
Cited by
-
Exposure to human relevant mixtures of halogenated persistent organic pollutants (POPs) alters neurodevelopmental processes in human neural stem cells undergoing differentiation.Reprod Toxicol. 2021 Mar;100:17-34. doi: 10.1016/j.reprotox.2020.12.013. Epub 2020 Dec 14. Reprod Toxicol. 2021. PMID: 33333158 Free PMC article.
-
Technical assessment of different extraction methods and transcriptome profiling of RNA isolated from small volumes of blood.Sci Rep. 2023 Mar 3;13(1):3598. doi: 10.1038/s41598-023-30629-5. Sci Rep. 2023. PMID: 36869090 Free PMC article.
-
Simultaneous extraction of mRNA and microRNA from whole blood stabilized in tempus tubes.BMC Res Notes. 2019 Jan 18;12(1):39. doi: 10.1186/s13104-019-4087-5. BMC Res Notes. 2019. PMID: 30658701 Free PMC article.
-
Restoration of Cognitive Performance in Mice Carrying a Deficient Allele of 8-Oxoguanine DNA Glycosylase by X-ray Irradiation.Neurotox Res. 2018 May;33(4):824-836. doi: 10.1007/s12640-017-9833-7. Epub 2017 Nov 3. Neurotox Res. 2018. PMID: 29101721
-
RNA from stabilized whole blood enables more comprehensive immune gene expression profiling compared to RNA from peripheral blood mononuclear cells.PLoS One. 2020 Jun 26;15(6):e0235413. doi: 10.1371/journal.pone.0235413. eCollection 2020. PLoS One. 2020. PMID: 32589655 Free PMC article.
References
-
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

