HIV-1 drug resistance and resistance testing
- PMID: 27587334
- PMCID: PMC5136505
- DOI: 10.1016/j.meegid.2016.08.031
HIV-1 drug resistance and resistance testing
Abstract
The global scale-up of antiretroviral (ARV) therapy (ART) has led to dramatic reductions in HIV-1 mortality and incidence. However, HIV drug resistance (HIVDR) poses a potential threat to the long-term success of ART and is emerging as a threat to the elimination of AIDS as a public health problem by 2030. In this review we describe the genetic mechanisms, epidemiology, and management of HIVDR at both individual and population levels across diverse economic and geographic settings. To describe the genetic mechanisms of HIVDR, we review the genetic barriers to resistance for the most commonly used ARVs and describe the extent of cross-resistance between them. To describe the epidemiology of HIVDR, we summarize the prevalence and patterns of transmitted drug resistance (TDR) and acquired drug resistance (ADR) in both high-income and low- and middle-income countries (LMICs). We also review to two categories of HIVDR with important public health relevance: (i) pre-treatment drug resistance (PDR), a World Health Organization-recommended HIVDR surveillance metric and (ii) and pre-exposure prophylaxis (PrEP)-related drug resistance, a type of ADR that can impact clinical outcomes if present at the time of treatment initiation. To summarize the implications of HIVDR for patient management, we review the role of genotypic resistance testing and treatment practices in both high-income and LMIC settings. In high-income countries where drug resistance testing is part of routine care, such an understanding can help clinicians prevent virological failure and accumulation of further HIVDR on an individual level by selecting the most efficacious regimens for their patients. Although there is reduced access to diagnostic testing and to many ARVs in LMIC, understanding the scientific basis and clinical implications of HIVDR is useful in all regions in order to shape appropriate surveillance, inform treatment algorithms, and manage difficult cases.
Keywords: Antiretroviral therapy; Diagnostic test; Drug resistance; HIV-1; Mutations; Surveillance.
Copyright © 2016 Elsevier B.V. All rights reserved.
Conflict of interest statement
Potential conflicts of interest: DSC receives research funding through the Bristol-Myers Squibb Virology Fellows Research Program. RWS is a consultant for Celera, and receives research funding from Roche Molecular, Gilead Sciences, Bristol-Myers Squibb, and Merck. The other authors have no potential conflicts of interest to disclose.
Figures



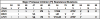
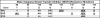
References
-
- Abram ME, Hluhanich RM, Goodman DD, Andreatta KN, Margot NA, Ye L, Niedziela-Majka A, Barnes TL, Novikov N, Chen X, Svarovskaia ES, McColl DJ, White KL, Miller MD. Impact of Primary Elvitegravir Resistance-Associated Mutations in HIV-1 Integrase on Drug Susceptibility and Viral Replication Fitness. Antimicrobial agents and chemotherapy 2013 - PMC - PubMed
-
- Antinori A, Zaccarelli M, Cingolani A, Forbici F, Rizzo MG, Trotta MP, Di Giambenedetto S, Narciso P, Ammassari A, Girardi E, De Luca A, Perno CF. Cross-resistance among nonnucleoside reverse transcriptase inhibitors limits recycling efavirenz after nevirapine failure. AIDS research and human retroviruses. 2002;18:835–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

