Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1
- PMID: 27587584
- PMCID: PMC5063001
- DOI: 10.1093/nar/gkw761
Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1
Abstract
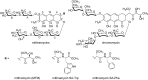
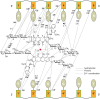
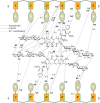
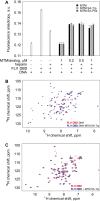
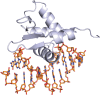
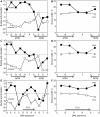
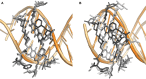
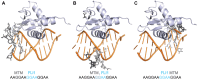
Transcription factors have been considered undruggable, but this paradigm has been recently challenged. DNA binding natural product mithramycin (MTM) is a potent antagonist of oncogenic transcription factor EWS-FLI1. Structural details of MTM recognition of DNA, including the FLI1 binding sequence GGA(A/T), are needed to understand how MTM interferes with EWS-FLI1. We report a crystal structure of an MTM analogue MTM SA-Trp bound to a DNA oligomer containing a site GGCC, and two structures of a novel analogue MTM SA-Phe in complex with DNA. MTM SA-Phe is bound to sites AGGG and GGGT on one DNA, and to AGGG and GGGA(T) (a FLI1 binding site) on the other, revealing how MTM recognizes different DNA sequences. Unexpectedly, at sub-micromolar concentrations MTMs stabilize FLI1-DNA complex on GGAA repeats, which are critical for the oncogenic function of EWS-FLI1. We also directly demonstrate by nuclear magnetic resonance formation of a ternary FLI1-DNA-MTM complex on a single GGAA FLI1/MTM binding site. These biochemical and structural data and a new FLI1-DNA structure suggest that MTM binds the minor groove and perturbs FLI1 bound nearby in the major groove. This ternary complex model may lead to development of novel MTM analogues that selectively target EWS-FLI1 or other oncogenic transcription factors, as anti-cancer therapeutics.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
The Position of Indole Methylation Controls the Structure, DNA Binding, and Cellular Functions of Mithramycin SA-Trp Analogues.Chembiochem. 2025 May 27;26(10):e202401084. doi: 10.1002/cbic.202401084. Epub 2025 May 6. Chembiochem. 2025. PMID: 40246689
-
Allosteric interference in oncogenic FLI1 and ERG transactions by mithramycins.Structure. 2021 May 6;29(5):404-412.e4. doi: 10.1016/j.str.2020.11.012. Epub 2020 Dec 3. Structure. 2021. PMID: 33275876 Free PMC article.
-
Solution structure of mithramycin dimers bound to partially overlapping sites on DNA.J Mol Biol. 1995 Sep 1;251(5):674-89. doi: 10.1006/jmbi.1995.0464. J Mol Biol. 1995. PMID: 7666419
-
The antitumor antibiotic mithramycin: new advanced approaches in modification and production.Appl Microbiol Biotechnol. 2020 Sep;104(18):7701-7721. doi: 10.1007/s00253-020-10782-x. Epub 2020 Jul 20. Appl Microbiol Biotechnol. 2020. PMID: 32686008 Review.
-
EWS-FLI1 in Ewing's sarcoma: real targets and collateral damage.Adv Exp Med Biol. 2006;587:41-52. doi: 10.1007/978-1-4020-5133-3_4. Adv Exp Med Biol. 2006. PMID: 17163154 Review.
Cited by
-
Chromomycin A2 potently inhibits glucose-stimulated insulin secretion from pancreatic β cells.J Gen Physiol. 2018 Dec 3;150(12):1747-1757. doi: 10.1085/jgp.201812177. Epub 2018 Oct 23. J Gen Physiol. 2018. PMID: 30352794 Free PMC article.
-
Mithramycin 2'-Oximes with Improved Selectivity, Pharmacokinetics, and Ewing Sarcoma Antitumor Efficacy.J Med Chem. 2020 Nov 25;63(22):14067-14086. doi: 10.1021/acs.jmedchem.0c01526. Epub 2020 Nov 16. J Med Chem. 2020. PMID: 33191745 Free PMC article.
-
A survey of recent unusual high-resolution DNA structures provoked by mismatches, repeats and ligand binding.Nucleic Acids Res. 2018 Jul 27;46(13):6416-6434. doi: 10.1093/nar/gky561. Nucleic Acids Res. 2018. PMID: 29945186 Free PMC article. Review.
-
Mithramycin and Analogs for Overcoming Cisplatin Resistance in Ovarian Cancer.Biomedicines. 2021 Jan 12;9(1):70. doi: 10.3390/biomedicines9010070. Biomedicines. 2021. PMID: 33445667 Free PMC article. Review.
-
The Position of Indole Methylation Controls the Structure, DNA Binding, and Cellular Functions of Mithramycin SA-Trp Analogues.Chembiochem. 2025 May 27;26(10):e202401084. doi: 10.1002/cbic.202401084. Epub 2025 May 6. Chembiochem. 2025. PMID: 40246689
References
-
- Delattre O., Zucman J., Plougastel B., Desmaze C., Melot T., Peter M., Kovar H., Joubert I., de Jong P., Rouleau G., et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. - PubMed
-
- May W.A., Gishizky M.L., Lessnick S.L., Lunsford L.B., Lewis B.C., Delattre O., Zucman J., Thomas G., Denny C.T. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5752–5756. - PMC - PubMed
-
- Arvand A., Welford S.M., Teitell M.A., Denny C.T. The COOH-terminal domain of FLI-1 is necessary for full tumorigenesis and transcriptional modulation by EWS/FLI-1. Cancer Res. 2001;61:5311–5317. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

