LuxGLM: a probabilistic covariate model for quantification of DNA methylation modifications with complex experimental designs
- PMID: 27587669
- PMCID: PMC5013920
- DOI: 10.1093/bioinformatics/btw468
LuxGLM: a probabilistic covariate model for quantification of DNA methylation modifications with complex experimental designs
Abstract
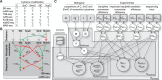
Motivation: 5-methylcytosine (5mC) is a widely studied epigenetic modification of DNA. The ten-eleven translocation (TET) dioxygenases oxidize 5mC into oxidized methylcytosines (oxi-mCs): 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). DNA methylation modifications have multiple functions. For example, 5mC is shown to be associated with diseases and oxi-mC species are reported to have a role in active DNA demethylation through 5mC oxidation and DNA repair, among others, but the detailed mechanisms are poorly understood. Bisulphite sequencing and its various derivatives can be used to gain information about all methylation modifications at single nucleotide resolution. Analysis of bisulphite based sequencing data is complicated due to the convoluted read-outs and experiment-specific variation in biochemistry. Moreover, statistical analysis is often complicated by various confounding effects. How to analyse 5mC and oxi-mC data sets with arbitrary and complex experimental designs is an open and important problem.
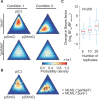
Results: We propose the first method to quantify oxi-mC species with arbitrary covariate structures from bisulphite based sequencing data. Our probabilistic modeling framework combines a previously proposed hierarchical generative model for oxi-mC-seq data and a general linear model component to account for confounding effects. We show that our method provides accurate methylation level estimates and accurate detection of differential methylation when compared with existing methods. Analysis of novel and published data gave insights into to the demethylation of the forkhead box P3 (Foxp3) locus during the induced T regulatory cell differentiation. We also demonstrate how our covariate model accurately predicts methylation levels of the Foxp3 locus. Collectively, LuxGLM method improves the analysis of DNA methylation modifications, particularly for oxi-mC species.
Availability and implementation: An implementation of the proposed method is available under MIT license at https://github.org/tare/LuxGLM/ CONTACT: taijo@simonsfoundation.org or harri.lahdesmaki@aalto.fi
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

