Thermal Unthreading of the Lasso Peptides Astexin-2 and Astexin-3
- PMID: 27588549
- PMCID: PMC5148663
- DOI: 10.1021/acschembio.6b00588
Thermal Unthreading of the Lasso Peptides Astexin-2 and Astexin-3
Abstract
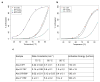
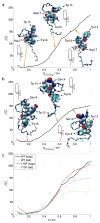
Lasso peptides are a class of knot-like polypeptides in which the C-terminal tail of the peptide threads through a ring formed by an isopeptide bond between the N-terminal amine group and a side chain carboxylic acid. The small size (∼20 amino acids) and simple topology of lasso peptides make them a good model system for studying the unthreading of entangled polypeptides, both with experiments and atomistic simulation. Here, we present an in-depth study of the thermal unthreading behavior of two lasso peptides astexin-2 and astexin-3. Quantitative kinetics and energetics of the unthreading process were determined for variants of these peptides using a series of chromatography and mass spectrometry experiments and biased molecular dynamics (MD) simulations. In addition, we show that the Tyr15Phe variant of astexin-3 unthreads via an unprecedented "tail pulling" mechanism. MD simulations on a model ring-thread system coupled with machine learning approaches also led to the discovery of physicochemical descriptors most important for peptide unthreading.
Figures






References
-
- Lim NCH, Jackson SE. Molecular knots in biology and chemistry. J Phys: Conds Matter. 2015;27 - PubMed
-
- Lim K, Zhang H, Tempczyk A, Krajewski W, Bonander N, Toedt J, Howard A, Eisenstein E, Herzberg O. Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): A cofactor bound at a site formed by a knot. Proteins: Struct Funct, Genet. 2003;51:56–67. - PubMed
-
- Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

