Evolution of New cis-Regulatory Motifs Required for Cell-Specific Gene Expression in Caenorhabditis
- PMID: 27588814
- PMCID: PMC5010242
- DOI: 10.1371/journal.pgen.1006278
Evolution of New cis-Regulatory Motifs Required for Cell-Specific Gene Expression in Caenorhabditis
Abstract
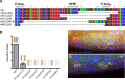
Patterning of C. elegans vulval cell fates relies on inductive signaling. In this induction event, a single cell, the gonadal anchor cell, secretes LIN-3/EGF and induces three out of six competent precursor cells to acquire a vulval fate. We previously showed that this developmental system is robust to a four-fold variation in lin-3/EGF genetic dose. Here using single-molecule FISH, we find that the mean level of expression of lin-3 in the anchor cell is remarkably conserved. No change in lin-3 expression level could be detected among C. elegans wild isolates and only a low level of change-less than 30%-in the Caenorhabditis genus and in Oscheius tipulae. In C. elegans, lin-3 expression in the anchor cell is known to require three transcription factor binding sites, specifically two E-boxes and a nuclear-hormone-receptor (NHR) binding site. Mutation of any of these three elements in C. elegans results in a dramatic decrease in lin-3 expression. Yet only a single E-box is found in the Drosophilae supergroup of Caenorhabditis species, including C. angaria, while the NHR-binding site likely only evolved at the base of the Elegans group. We find that a transgene from C. angaria bearing a single E-box is sufficient for normal expression in C. elegans. Even a short 58 bp cis-regulatory fragment from C. angaria with this single E-box is able to replace the three transcription factor binding sites at the endogenous C. elegans lin-3 locus, resulting in the wild-type expression level. Thus, regulatory evolution occurring in cis within a 58 bp lin-3 fragment, results in a strict requirement for the NHR binding site and a second E-box in C. elegans. This single-cell, single-molecule, quantitative and functional evo-devo study demonstrates that conserved expression levels can hide extensive change in cis-regulatory site requirements and highlights the evolution of new cis-regulatory elements required for cell-specific gene expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

