Substrate Specificity of SAMHD1 Triphosphohydrolase Activity Is Controlled by Deoxyribonucleoside Triphosphates and Phosphorylation at Thr592
- PMID: 27588835
- PMCID: PMC5225984
- DOI: 10.1021/acs.biochem.6b00627
Substrate Specificity of SAMHD1 Triphosphohydrolase Activity Is Controlled by Deoxyribonucleoside Triphosphates and Phosphorylation at Thr592
Abstract
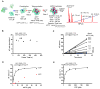
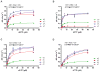
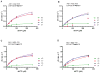
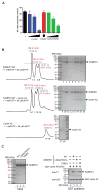
The sterile alpha motif (SAM) and histidine-aspartate (HD) domain containing protein 1 (SAMHD1) constitute a triphosphohydrolase that converts deoxyribonucleoside triphosphates (dNTPs) into deoxyribonucleosides and triphosphates. SAMHD1 exists in multiple states. The monomer and apo- or GTP-bound dimer are catalytically inactive. Binding of dNTP at allosteric site 2 (AS2), adjacent to GTP-binding allosteric site 1 (AS1), induces formation of the tetramer, the catalytically active form. We have developed an enzyme kinetic assay, tailored to control specific dNTP binding at each site, allowing us to determine the kinetic binding parameters of individual dNTPs at both the AS2 and catalytic sites for all possible combinations of dNTP binding at both sites. Here, we show that the apparent Km values of dNTPs at AS2 vary in the order of dCTP < dGTP < dATP < dTTP. Interestingly, dCTP binding at AS2 significantly reduces the dCTP hydrolysis rate, which is restored to a rate comparable to that of other dNTPs upon dGTP, dATP, or dTTP binding at AS2. Strikingly, a phosphomimetic mutant, Thr592Asp SAMHD1 as well as phospho-Thr592, show a significantly altered substrate specificity, with the rate of dCTP hydrolysis being selectively reduced regardless of which dNTP binds at AS2. Furthermore, cyclin A2 binding at the C-terminus of SAMHD1 induces the disassembly of the SAMHD1 tetramer, suggesting an additional layer of SAMHD1 activity modulation by cyclin A2/CDK2 kinase. Together, our results reveal multiple allosteric mechanisms for controlling the rate of dNTP destruction by SAMHD1.
Figures


References
-
- Hakansson P, Dahl L, Chilkova O, Domkin V, Thelander L. The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribonucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J Biol Chem. 2006;281:1778–1783. - PubMed
-
- Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281:7834–7841. - PubMed
-
- Bebenek K, Roberts JD, Kunkel TA. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J Biol Chem. 1992;267:3589–3596. - PubMed
-
- Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

