Population genomics identifies the origin and signatures of selection of Korean weedy rice
- PMID: 27589078
- PMCID: PMC5316921
- DOI: 10.1111/pbi.12630
Population genomics identifies the origin and signatures of selection of Korean weedy rice
Abstract
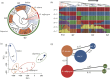
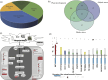
Weedy rice is the same biological species as cultivated rice (Oryza sativa); it is also a noxious weed infesting rice fields worldwide. Its formation and population-selective or -adaptive signatures are poorly understood. In this study, we investigated the phylogenetics, population structure and signatures of selection of Korean weedy rice by determining the whole genomes of 30 weedy rice, 30 landrace rice and ten wild rice samples. The phylogenetic tree and results of ancestry inference study clearly showed that the genetic distance of Korean weedy rice was far from the wild rice and near with cultivated rice. Furthermore, 537 genes showed evidence of recent positive or divergent selection, consistent with some adaptive traits. This study indicates that Korean weedy rice originated from hybridization of modern indica/indica or japonica/japonica rather than wild rice. Moreover, weedy rice is not only a notorious weed in rice fields, but also contains many untapped valuable traits or haplotypes that may be a useful genetic resource for improving cultivated rice.
Keywords: genetic resources; natural selection; origin; weedy rice; whole-genome resequencing.
© 2016 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures

References
-
- Bres‐Patry, C. , Bangratz, M. and Ghesquiere, A. (2001) Genetic diversity and population dynamics of weedy rice in Camargue area [France]. Genet. Sel. Evol. 33, 425–440.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

