Normalizing Microbiota-Induced Retinoic Acid Deficiency Stimulates Protective CD8(+) T Cell-Mediated Immunity in Colorectal Cancer
- PMID: 27590114
- PMCID: PMC5132405
- DOI: 10.1016/j.immuni.2016.08.008
Normalizing Microbiota-Induced Retinoic Acid Deficiency Stimulates Protective CD8(+) T Cell-Mediated Immunity in Colorectal Cancer
Abstract
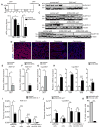
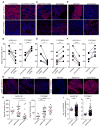
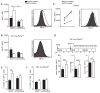
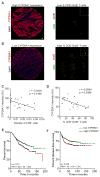
Although all-trans-retinoic acid (atRA) is a key regulator of intestinal immunity, its role in colorectal cancer (CRC) is unknown. We found that mice with colitis-associated CRC had a marked deficiency in colonic atRA due to alterations in atRA metabolism mediated by microbiota-induced intestinal inflammation. Human ulcerative colitis (UC), UC-associated CRC, and sporadic CRC specimens have similar alterations in atRA metabolic enzymes, consistent with reduced colonic atRA. Inhibition of atRA signaling promoted tumorigenesis, whereas atRA supplementation reduced tumor burden. The benefit of atRA treatment was mediated by cytotoxic CD8(+) T cells, which were activated due to MHCI upregulation on tumor cells. Consistent with these findings, increased colonic expression of the atRA-catabolizing enzyme, CYP26A1, correlated with reduced frequencies of tumoral cytotoxic CD8(+) T cells and with worse disease prognosis in human CRC. These results reveal a mechanism by which microbiota drive colon carcinogenesis and highlight atRA metabolism as a therapeutic target for CRC.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
-
- Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. Journal of lipid research. 2002;43:1773–1808. - PubMed
-
- Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, Moriyama EH, Copeland JK, Kumar S, Green B, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288–299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

