Decoding the Long Noncoding RNA During Cardiac Maturation: A Roadmap for Functional Discovery
- PMID: 27591185
- PMCID: PMC5085833
- DOI: 10.1161/CIRCGENETICS.115.001363
Decoding the Long Noncoding RNA During Cardiac Maturation: A Roadmap for Functional Discovery
Abstract
Background: Cardiac maturation during perinatal transition of heart is critical for functional adaptation to hemodynamic load and nutrient environment. Perturbation in this process has major implications in congenital heart defects. Transcriptome programming during perinatal stages is an important information but incomplete in current literature, particularly, the expression profiles of the long noncoding RNAs (lncRNAs) are not fully elucidated.
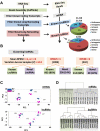
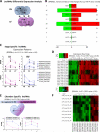
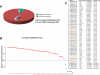
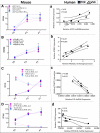
Methods and results: From comprehensive analysis of transcriptomes derived from neonatal mouse heart left and right ventricles, a total of 45 167 unique transcripts were identified, including 21 916 known and 2033 novel lncRNAs. Among these lncRNAs, 196 exhibited significant dynamic regulation along maturation process. By implementing parallel weighted gene co-expression network analysis of mRNA and lncRNA data sets, several lncRNA modules coordinately expressed in a developmental manner similar to protein coding genes, while few lncRNAs revealed chamber-specific patterns. Out of 2262 lncRNAs located within 50 kb of protein coding genes, 5% significantly correlate with the expression of their neighboring genes. The impact of Ppp1r1b-lncRNA on the corresponding partner gene Tcap was validated in cultured myoblasts. This concordant regulation was also conserved in human infantile hearts. Furthermore, the Ppp1r1b-lncRNA/Tcap expression ratio was identified as a molecular signature that differentiated congenital heart defect phenotypes.
Conclusions: The study provides the first high-resolution landscape on neonatal cardiac lncRNAs and reveals their potential interaction with mRNA transcriptome during cardiac maturation. Ppp1r1b-lncRNA was identified as a regulator of Tcap expression, with dynamic interaction in postnatal cardiac development and congenital heart defects.
Keywords: congenital cardiac defect; gene regulation; lncRNA; neonatal heart maturation; neonatal mouse cardiomyocyte; transcriptome.
© 2016 American Heart Association, Inc.
Figures



Comment in
-
Of Caps and Gaps, Postnatal Hearts, Elusive Facts, and lncs.Circ Cardiovasc Genet. 2016 Oct;9(5):389-391. doi: 10.1161/CIRCGENETICS.116.001608. Circ Cardiovasc Genet. 2016. PMID: 27756779 No abstract available.
References
-
- Finnemore A, Groves A. Physiology of the fetal and transitional circulation. Semin Fetal Neonatal Med. 2015;20:210–216. - PubMed
-
- Donn MS. Fetal-to-neonatal maladaptation. Semin Fetal Neonatal Med. 2006;11:166–173. - PubMed
-
- Harvey RP. Patterning the vertebrate heart. Nat Rev Genet. 2002;3:544–556. - PubMed
-
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006b;126:1037–1048. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

