Congenital Heart Disease Genetics Uncovers Context-Dependent Organization and Function of Nucleoporins at Cilia
- PMID: 27593162
- PMCID: PMC5021619
- DOI: 10.1016/j.devcel.2016.08.002
Congenital Heart Disease Genetics Uncovers Context-Dependent Organization and Function of Nucleoporins at Cilia
Abstract
Human genomics is identifying candidate genes for congenital heart disease (CHD), but discovering the underlying mechanisms remains challenging. In a patient with CHD and heterotaxy (Htx), a disorder of left-right patterning, we previously identified a duplication in Nup188. However, a mechanism to explain how a component of the nuclear pore complex (NPC) could cause Htx/CHD was undefined. Here, we show that knockdown of Nup188 or its binding partner Nup93 leads to a loss of cilia during embryonic development while leaving NPC function largely intact. Many data, including the localization of endogenous Nup188/93 at cilia bases, support their direct role at cilia. Super-resolution imaging of Nup188 shows two barrel-like structures with dimensions and organization incompatible with an NPC-like ring, arguing against a proposed "ciliary pore complex." We suggest that the nanoscale organization and function of nucleoporins are context dependent in a way that is required for the structure of the heart.
Keywords: 3D nanoscopy; Xenopus tropicalis; congenital heart disease; heterotaxy; left-right patterning; nuclear pore complex; nucleoporin; super-resolution.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
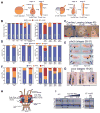
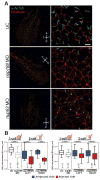
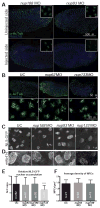
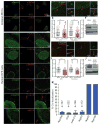

Comment in
-
Cilia and Nuclear Pore Proteins: Pore No More?Dev Cell. 2016 Sep 12;38(5):445-6. doi: 10.1016/j.devcel.2016.08.019. Dev Cell. 2016. PMID: 27623377
References
-
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
-
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. - PubMed
-
- Basu B, Brueckner M. Cilia multifunctional organelles at the center of vertebrate left-right asymmetry. Curr Top Dev Biol. 2008;85:151–174. - PubMed
-
- Bisgrove BW, Yost HJ. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

