MultiSyn: A Webtool for Multiple Synteny Detection and Visualization of User's Sequence of Interest Compared to Public Plant Species
- PMID: 27594782
- PMCID: PMC5003123
- DOI: 10.4137/EBO.S40009
MultiSyn: A Webtool for Multiple Synteny Detection and Visualization of User's Sequence of Interest Compared to Public Plant Species
Abstract
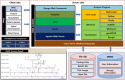
Information on multiple synteny between plants and/or within a plant is key information to understand genome evolution. In addition, visualization of multiple synteny is helpful in interpreting evolution. So far, some web applications have been developed to determine and visualize multiple homology regions at once. However, the applications are not fully convenient for biologists because some of them do not include the function of synteny determination but visualize the multiple synteny plots by allowing users to upload their synteny data by determining the synteny based only on BLAST similarity information, with some algorithms not designed for synteny determination. Here, we introduce a web application that determines and visualizes multiple synteny from two types of files, simplified browser extensible data and protein sequence file by MCScanX algorithm, which have been used in many synteny studies.
Keywords: detection; multiple synteny; plant; visualization; webtool.
Figures


References
-
- Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20(18):3643–6. - PubMed
-
- Simillion C, Janssens K, Sterck L, Van de Peer Y. i-ADHoRe 2.0: an improved tool to detect degenerated genomic homology using genomic profiles. Bioinformatics. 2008;24:127–8. - PubMed
-
- Zeng X, Pei J, Vergara IA, Nesbitt MJ, Wang K, Chen N. EDBT. Nantes, France: 2008. OrthoCluster: a new tool for mining synteny blocks and applications in comparative genomics.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

